Fig 1.
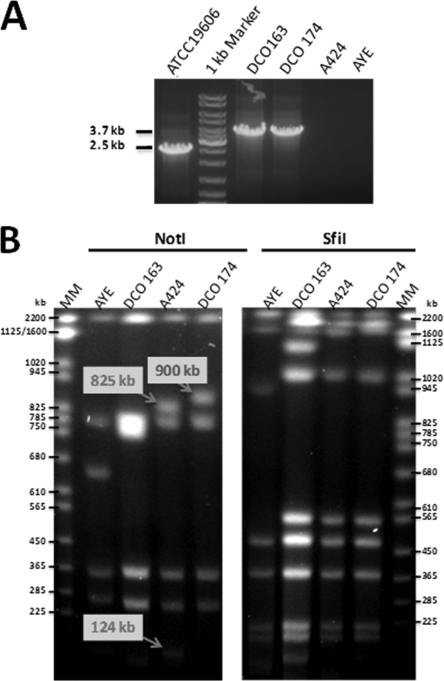
Analysis of the putative TnAbaR23-minus mutants of Acinetobacter baumannii A424. (A) PCR amplification across the targeted deleted locus in DCO163 and DCO174 mutant strains using the primers AbaRUF2 and AbaRDR2 yielded the expected ∼3.7-kb product, while failing to amplify across the A424 TnAbaR23 element. The sequenced strains ATCC 19606 and AYE that lacked and harbored an island within the comM gene were used as positive (2.5-kb band expected) and negative PCR controls, respectively. (B) Pulsed-field gel electrophoresis analysis of NotI- and SfiI-digested A424, DCO163, DCO174, and AYE high-molecular-weight genomic DNA. The three bands with arrows and labels indicate the only observed NotI profile differences between A424 and DCO174. Subsequently available sequence data showed that TnAbaR23 harbors two very closely spaced NotI sites, while the replacement cassette lacks a NotI site. Hence, the observed loss of two A424 bands (825 kb plus 124 kb) and the appearance of a new DCO174-specific band (900 kb), with the difference between the two being the size of TnAbaR23 (48.3 kb). Lanes MM contain molecular size markers.