Abstract
The taxonomic class of oomycetes contains numerous pathogens of plants and animals but is related to nonpathogenic diatoms and brown algae. Oomycetes have flexible genomes comprising large gene families that play roles in pathogenicity. The evolutionary processes that shaped the gene content have not yet been studied by applying systematic tree reconciliation of the phylome of these species. We analyzed evolutionary dynamics of ten Stramenopiles. Gene gains, duplications, and losses were inferred by tree reconciliation of 18,459 gene trees constituting the phylome with a highly supported species phylogeny. We reconstructed a strikingly large last common ancestor of the Stramenopiles that contained ∼10,000 genes. Throughout evolution, the genomes of pathogenic oomycetes have constantly gained and lost genes, though gene gains through duplications outnumber the losses. The branch leading to the plant pathogenic Phytophthora genus was identified as a major transition point characterized by increased frequency of duplication events that has likely driven the speciation within this genus. Large gene families encoding different classes of enzymes associated with pathogenicity such as glycoside hydrolases are formed by complex and distinct patterns of duplications and losses leading to their expansion in extant oomycetes. This study unveils the large-scale evolutionary dynamics that shaped the genomes of pathogenic oomycetes. By the application of phylogenetic based analyses methods, it provides additional insights that shed light on the complex history of oomycete genome evolution and the emergence of large gene families characteristic for this important class of pathogens.
Keywords: oomycetes, genome reconstruction, evolution, gene families, evolutionary dynamics, tree reconciliation
Introduction
Recent comparative genome analyses of Stramenopiles have facilitated initial insights into the evolution and lifestyle of the individual species within this lineage and in particular of pathogenic oomycetes (Tyler et al. 2006; Martens et al. 2008; Haas et al. 2009; Gobler et al. 2011; Seidl et al. 2011). The extensive Stramenopile lineage comprises species that cover diverse ecological niches and lifestyles ranging from photosynthetic diatoms and brown algae to filamentous heterotrophic oomycetes. According to the controversial Chromalveolate hypothesis, Stramenopiles are grouped together with other chlorophyll-c containing lineages such as Crypophytes, Alveolates, and Haptophytes into one monophyletic supergroup (Cavalier-Smith 1999; Keeling 2009), sometimes also referred to as CASH. This grouping has been rationalized on the hypothesis that the last common ancestor (LCA) of these lineages acquired its plastid from a single initial event of secondary endosymbiosis with a red alga that has been subsequently inherited strictly vertically. Consequently, plastid-lacking species within CASH lineages have lost their plastids secondarily and independently. The competing serial eukaryotic–-eukaryotic endosymbiotic (SEEE) hypothesis proposes an independent spread of plastids within CASH lineages, and hence, dependent on the time point of acquisition, no secondary losses are needed to explain the lack of plastids in several taxa throughout all lineages (Cavalier-Smith et al. 1994; Archibald 2009; Baurain et al. 2010).
The plastid-lacking oomycetes are saprophytes or pathogens of plants and animals with huge economical as well as ecological impact (Govers and Gijzen 2006). Well known are the notorious late blight pathogen Phytophthora infestans that infects both tomato and potato and the animal pathogen Saprolegnia parasitica that causes saprolegniasis, for example, in salmon. Within the oomycetes studied so far, the genomes of Phytophthora spp. have by far the largest genomes, ranging from 65 up to 240 Mb (supplementary additional file 1A, Supplementary Material online). This broad variation in genome sizes is also observed among fungi, many of which are pathogens that exploit infection strategies similar to oomycetes (Latijnhouwers et al. 2003). Within Ascomycetes, for example, the rice blast fungus Magnaporthe grisea has a relatively small genome (38 Mb, ∼12,000 predicted genes), whereas the recently sequenced genome of the obligate biotrophic powdery mildew fungus Blumeria graminis is considerably larger (∼100 Mb); the expansion is mainly caused by transposable elements (Spanu et al. 2010; Duplessis et al. 2011). It has been speculated that Phytophthora spp. might have undergone a whole-genome duplication or at least several large-scale duplications. That, together with their divergent repertoire of transposable elements, probably contributed to the increased genome size and gene content of the Phytophthora spp. (Jiang et al. 2005; Haas et al. 2009; Martens and Van de Peer 2010).
Oomycete pathogens have a large and diverse repertoire of expanded gene families (Tyler et al. 2006; Haas et al. 2009; Baxter et al. 2010; Lévesque et al. 2010; Seidl et al. 2011). These mainly encode proteins that are secreted and implied to be directly or indirectly involved in pathogenicity, such as the NEP1-like proteins (Gijzen and Nürnberger 2006) or glycoside hydrolases (Ospina-Giraldo et al. 2010; Seidl et al. 2011). Two notable classes of highly abundant genes that are identified in several oomycete genomes encode secreted proteins characterized by the presence of either the RXLR or the LXLFLAK (Crinkler) motif (Whisson et al. 2007; Dou et al. 2008; Jiang et al. 2008; Haas et al. 2009). These motifs, located in the N-terminal region of the mature protein, play a role in translocation of the proteins from the apoplast to the cytoplasm of the host cell; however, the process is not yet fully understood (Govers and Bouwmeester 2008; Kale et al. 2010; Stassen and Van den Ackerveken 2011).
Initial analyses of the evolution of several pathogenic oomycetes led to the identification of large gene families. However, the individual contributions and the exact sequence of different evolutionary processes such as gene gains, duplications, and losses that caused the enormous increase in gene families sizes are still unknown. We studied these dynamics, and also the general evolution of the gene content, by a phylogenetic approach that reconciled 18,459 individual gene trees that constitute the phylome of Stramenopiles with a reliable species phylogeny. This systematic and comprehensive analysis of the evolutionary events is now feasible because several genomes of oomycetes and their sister lineages have been sequenced, a substantial increase to previous studies. We have utilized the predicted proteomes of six pathogenic oomycetes and four nonpathogenic Stramenochromes (supplementary additional file 2, Supplementary Material online), a sublineage within the Stramenopiles (Patterson 1999). The six oomycetes comprise the fish pathogen S. parasitica and five plant pathogens: the necrotrophic wide host range pathogen Pythium ultimum, the obligate downy mildew pathogen of Arabidopsis Hyaloperonospora arabidopsidis, and three Phytophthora species, P. infestans, P. sojae, and P. ramorum. The latter two cause stem and root rot on soybean and sudden oak death, respectively. The four aquatic photosynthetic Stramenochromes include the brown alga Ectocarpus siliculosus, the golden-brown alga Aureococcus anophagefferens, and two diatoms: Phaeodactylum tricornutum and Thalassiosira pseudonana. Our phylogeny-based approach resulted in an overview of the fundamental evolutionary dynamics underlying major transition points in the evolution of pathogenic oomycetes and how these differences are reflected in the expansion and contraction pattern of distinct functional classes, such as transcription regulation or carbohydrate metabolism. Moreover, we were able to elucidate the evolutionary history of large gene families in oomycetes, such as glycoside hydrolases and peptidases. These families show distinct evolutionary trajectories that caused their abundance in extant taxa, an observation that would not have been possible solely on parsimony- or abundance-based methods. This, together with our other results, highlights the needs for an advanced phylogeny-based analysis of the expansion of large gene families in the future.
Materials and Methods
To define protein families in the ten analyzed Stramenopiles, we created a sparse network based on Blast (Altschul et al. 1990) all-versus-all sequence similarity search (e value cutoff: 1 × 10−3). Spurious connections between short segments of similarity were removed, and the network was portioned into families using the Markov clustering algorithm (Van Dongen 2000; Enright et al. 2002). The presence of transposable elements in the proteomes was predicted by two independent methods and families containing at least one identified transposable element was removed.
A maximum likelihood phylogenetic tree was inferred using RAxML (Stamatakis 2006) (v7.0.4) with a gamma model of heterogeneity and Whelan and Goldman amino acid substitution matrix. A phylogenetic marker was created by concatenation of individual alignments of single-copy families derived by mafft (Katoh et al. 2002) (L-INS-I algorithm). The robustness of the topology was assessed by 1,000 bootstrap replicates. Relative divergence times within the Stramenopiles were estimated with BEAST under a strict clock model (Drummond and Rambaut 2007). The age prior for the last Stramenopile common ancestor (LSCA) was arbitrarily set to 100. We ran ten independent chains with 4,000,000 generations and subsequently averaged the estimates on the relative divergence times. The probability of the deviation between the observed and the expected number of evolutionary events at each branch was assessed by Poisson distribution.
We aligned the individual protein families, subsequently constructed RAxML maximum likelihood trees and assessed the robustness of these with 100 bootstrap replicates. We used NOTUNG (Chen et al. 2000; Durand et al. 2006) (v2.6; 1.5 duplication and 1 loss cost) to reconcile these trees with the species phylogeny. Uncertainties in the protein tree topology were assessed and weakly supported branches (<80% bootstrap support) were rearranged to minimize duplication/loss costs. Orthologous groups (OGs) were formed based on duplications at the LSCA. Consequently, each OG represents a single gene at LSCA or at the point of gain. All OGs are deposited under http://bioinformatics.bio.uu.nl/michael/index_supplementary.html.
Individual OGs were functionally annotated by transfer of clusters of orthologous groups (COG) classification from eggNOG (Muller et al. 2010), by functional annotation of chloroplast-associated proteins via gene ontology utilizing Blast2GO (Conesa et al. 2005), and by prediction of secretion signals and/or of host-cell translocation motifs (RxLR/LXLFLAK) or based on differential expression of the encoding genes during infection of the host. The prediction of signature Pfam domains identified OGs containing glycoside hydrolases and peptidases. Significant over- or underrepresentation of evolutionary events was assessed using Fisher’s exact test, and multiple testing correction was applied.
Complete information regarding all methods and material used for the analyses are reported in supplementary additional file 2 (Supplementary Material online).
Results
Protein Family Assignment
To systematically study the evolutionary dynamics of protein families in ten Stramenopile species, we classified the combined set of 148,744 predicted proteins into families (Materials and Methods). In total, 18,979 families were formed, and for 27,342 single sequences (singletons), no homology could be established.
Filtering for transposable elements resulted in the removal of 7,905 proteins representing 519 families and 267 singletons. Stramenopiles, in particular oomycetes and the brown alga E. siliculosus, contain a large and diverse repertoire of transposable elements (Jiang et al. 2005; Tyler et al. 2006; Haas et al. 2009; Cock et al. 2010). Relics of those have been observed in high abundance in the predicted proteomes and would have biased our analysis (Seidl et al. 2011). In total, this resulted in 45,535 families including 27,075 singletons (supplementary additional file 3A, Supplementary Material online). Other large-scale studies conducted in closely related phyla revealed a comparable number of singletons per genome (supplementary additional file 3B, Supplementary Material online) (see e.g., Martens et al. 2008; Cock et al. 2010). However, a direct comparison is not feasible because different species sets were used in the other studies. The remaining 18,459 multisequence families were used for tree reconciliation.
Species Phylogeny Utilizing Concatenated Single-Copy Genes
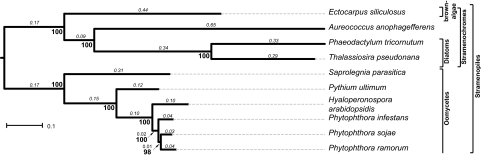
The quality of tree reconciliation is highly dependent on a correct species phylogeny. Furthermore, individual gene trees do not necessarily reflect the true relationship between species. In order to elucidate a reliable species phylogeny, we concatenated multiple families of single-copy genes, that is, families with only one member in each of the ten species included in this study (fig. 1). We concatenated alignments of 189 single-copy families and inferred the species phylogeny using a maximum likelihood approach implemented in RAxML (Stamatakis 2006). The robustness was assessed by 1,000 bootstrap replicates. The obtained species phylogeny is highly supported with bootstrap values >95% for all nodes. It mostly resembles the known topology of the tree of life, clearly separating the pathogenic oomycetes from the nonpathogenic Stramenochromes. However, the exact relationships within the genus Peronosporales contradict previous studies that either grouped P. sojae and P. infestans (Blair et al. 2008) or proposed the paraphyly of Phytophthora by grouping P. infestans as a sister taxa to H. arabidopsidis (Runge et al. 2011). Our phylogenetic analysis revealed a closer relationship between P. ramorum and P. sojae, and we show that this topology is more parsimonious in reconciliation of evolutionary events; hence, it was used for all further analyses (supplementary additional file 4A, Supplementary Material online).
FIG. 1.—
Maximum likelihood phylogeny of the analyzed Stramenopiles based on 189 concatenated marker families (branch lengths in “substitutions per site” are displayed in italics). The robustness of the topology was assessed using 1,000 bootstrap replicates (bold numbers).
Systematic Tree Reconciliation Guides Genome Reconstruction
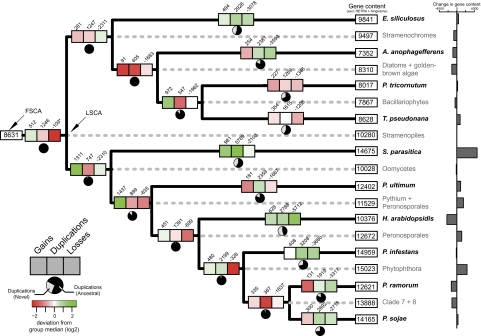
We obtained a comprehensive and dynamic picture of Stramenopile genome evolution by projecting gene gains, duplications, as well as losses onto the species phylogeny (fig. 2). For each of the 18,459 families, we inferred maximum likelihood trees, reconciled these with the predicted species phylogeny of Stramenopiles, and subsequently formed 19,596 OGs that represent single genes either at the LSCA or at the respective point of gain of individual OGs. To appropriately describe the evolutionary events that can affect an OG, these groups can also contain genes that descend from a duplication event subsequent to the reference speciation event (LSCA in our case), so called in-paralogs: these genes are related to the other genes within the group with respect to the reference speciation event (LSCA) and are hence orthologous (Fitch 2000; Sonnhammer and Koonin 2002). Consequently, an OG can reflect single-copy orthologs, but also more complex 1:n, n:m relationships, and is used as such throughout the manuscript.
FIG. 2.—
Projected evolutionary events on the Stramenopile phylogeny. The number of evolutionary events, that is, gene gains, duplications, and losses, are projected onto each branch of the phylogeny. Pie charts indicate the relative contribution of novel or ancestral OGs to the total number of duplications. The heat map highlights the deviation of the number of events from the median of the class (gains, duplications, or losses). Predicted gene content of the ancestors, LSCA and first Stramenopile common ancestor, as well as of the extant taxa (excluding singletons and transposable elements) is displayed in terminal boxes, whereas the calculated change in gene content, that is, change in the number of genes per branch, is shown by bar charts.
Over 50% of OGs are present in the LSCA. We found homologs outside of Stramenopiles for 95% (∼9,750) of these groups, and hence, they predate the LSCA. Based on our data set, the reconstructed genome of the LSCA contained at least 10,280 genes and is consequently remarkably large compared with the genome content of the Stramenochromes. The genes present in the LSCA are enriched for basic cellular functions, like transcription and translation. It is striking to see that of the remaining gains, 30% is observed at the LCA of oomycetes and the LCA of the Pythium + Peronosporales clade (1,311 and 1,437, respectively); the highest number of gene gain observed at any branch (P value < 0.01, Wilcoxon rank sum test [one-sided]). This demonstrates that gains, accompanied by duplications, have caused the increase in genome content of pathogenic oomycetes.
Despite the fact that Stramenochromes, unlike pathogenic oomycetes, show only small net changes in the number of encoded proteins (fig. 2), their genomes are not static. Similar to oomycete genomes, they are in constant flux: High numbers of duplications are balanced by an equally high number of losses. The contribution of individual duplications and losses on the same branch and the effect on the size of the OG could not have been observed with parsimony-based methods because many of these duplications and losses occur in the same OG on the same branch. Globally, we observed an average of 1.77 duplication and 2.06 losses per OG; however, only few OGs contribute to the majority of evolutionary events (e.g., members of the major facilitator superfamily or amino acid transporters).
To assess whether the observed duplication or loss events per individual branch deviate from the expected number, we calculated the relative frequency of these events per branch. Hence, we inferred branch length by estimating the relative divergence time of Stramenopiles using BEAST (Drummond and Rambaut 2007) and artificially dating the LSCA to 100 units of time (supplementary additional file 4B, Supplementary Material online). We predicted the position of the root by adding the ciliate Paramecium tetraurelia as an outgroup species. Based on the cumulative branch lengths (supplementary additional file 4B, Supplementary Material online) and the duplication and loss events (fig. 2), we estimated the relative frequency of duplications and losses to be 67 and 78 per unit of time, respectively. We contrasted the observed number of duplications/losses with expected numbers based on the global frequency and the length of the individual branch. The probability that the observed events deviate from the expectations was calculated using Poisson distribution. The abundance of observed duplications and losses significantly deviate from the expected number of events at each branch (supplementary additional file 5, Supplementary Material online). Within the Peronosporales clade, duplications and losses are significantly higher than expected (supplementary additional file 5, Supplementary Material online; duplications up to a maximum of ∼7-fold; 2.83 log2 fold), indicating an increased turnover of gene families in this clade. Interestingly, also at the LCA of Stramenochromes as well as the LCA of diatoms/golden-brown algae, the abundance of losses is significantly higher than expected, pointing to the contraction of OGs within the Stramenochromes.
A notable example of genome contraction is observed in the downy mildew Hyaloperonospora arabidopsidis. An accumulation of losses is accompanied by a lower number of duplication events. It is the only branch in the phylogeny where the majority of duplications occurs in lineage-specific groups. Hence, the H. arabidopsidis genome encodes a unique repertoire of expanded OGs, while at the same time, ancestral OGs, that is, OGs that were already gained before the point of duplications, were either completely lost or contracted in size.
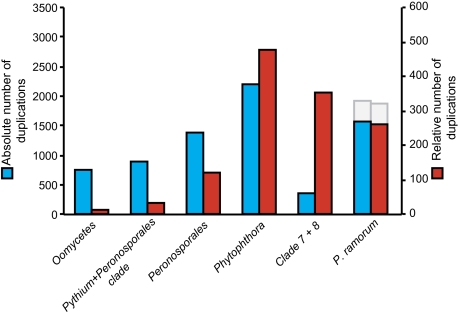
The increased genome content of the extant oomycetes is mainly caused by three events: gains, continuous duplications at internal branches of the species phylogeny, and a high number of duplications at branches leading to the extant taxa, for example, P. infestans, S. parasitica, and P. ultimum. Duplications at the LCAs are in general of lower abundance and affect ancestral OGs. A notable exception is the observed accumulation of duplications at the LCA of Phytophthora spp. (2.83-fold (log2) higher than expected) (fig. 2; supplementary additional file 5, Supplementary Material online); this is 1.5 times higher compared with the other duplications at internal branches. The increased number of duplications is even more pronounced when considering the relative number of duplication events per branch instead of the absolute abundance and hence points to a major duplication event in the evolution of the Phytophthora genus (fig. 3 and supplementary additional file 6, Supplementary Material online).
FIG. 3.—
Absolute and relative numbers of duplication events for P. ramorum and all its ancestors. The absolute number of duplications is displayed in blue, whereas the relative number of duplications (per unit of time) is shown in red. The gray bar represents the abundance of duplications (absolute and relative) including duplications occurring in lineage-specific OGs in P. ramorum.
Differences in the Evolutionary Dynamics of Biologically Distinct Functional Classes
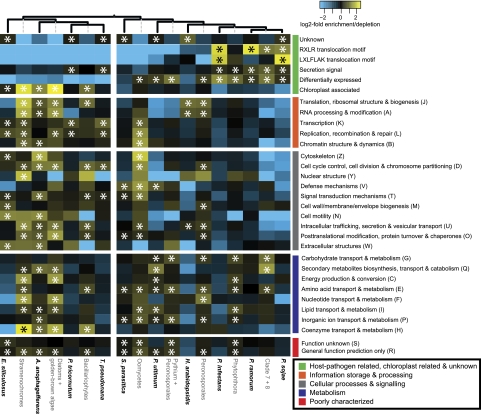
OGs can be assigned to functional classes by projecting the biological function of its individual proteins to the entire OG. We formed broad classes of functionally related OGs by transferring functional annotations from homologs based on the COG functional classification schema (Tatusov et al. 1997) and from predictions, for example, signal peptides or host cell translocation motifs (RXLR and LXLFLAK) (supplementary additional file 2, Supplementary Material online). These broad functional classes behave strikingly different with respect to their evolutionary pattern of expansion (duplications) and contraction (losses): They are either significantly overrepresented or significantly underrepresented at various points in the evolution of Stramenopiles (fig. 4 and supplementary additional file 7, Supplementary Material online).
Fig. 4.—
Differences in evolutionary trajectories between distinct functional classes. Over- or underrepresentation of duplication events at distinct branches of the species phylogeny observed for different functional classes (abbreviation for COG classes are displayed behind the description). The heat map shows the fold (log2) enrichment/depletion in duplications (saturating at −2 and 2). Significance of the overrepresentation/underrepresentation was assessed using a Fisher's exact test (P < 0.05), and multiple testing correction was addressed using a false discovery rate (q < 0.05). Significant enrichment is indicated by *; for both significant enrichment and depletion, see also supplementary additional file 7A (Supplementary Material online).
Overall, OGs belonging to COG “information processing and storage” or “cellular processes and signaling” have significantly more duplications at the LCA of oomycetes and within the Stramenochromes than at other branches. In contrast, OGs implied in host–pathogen interaction such as functional classes that contain RXLR and LXLFLAK motifs, secretion signals, as well as genes differentially expressed during infection of the host, predominantly expand within pathogenic oomycetes, both on internal as well as external branches. OGs containing predicted secreted proteins significantly expand at the LCA of Phytophthora spp. and throughout the genus, even though the analyzed Stramenopiles do not differ in absolute and relative size of the predicted secretomes (supplementary additional file 1B, Supplementary Material online).
Pathogenicity is not the only characteristic that discriminates the analyzed oomycetes and Stramenochromes because Stramenochromes are plastid-harboring photosynthetic active organisms. This lifestyle difference is clearly reflected in the observed evolutionary pattern of OGs containing proteins with functional association to the chloroplast (fig. 4; supplementary additional file 8, Supplementary Material online). Like the pathogenicity related OGs, these OGs are also highly dynamic in their evolution: They significantly expand at the LCAs within the Stramenochromes as well as at the branch leading to A. anophagefferens and significantly contract at the terminal branches and at the LCA of the diatoms. Interestingly, even though oomycetes do not harbor any plastids, we observed a considerable number of genes within the oomycete genomes that belong to ∼450 different chloroplast-associated OGs (supplementary additional file 8, Supplementary Material online). At the same time, as expected, losses of chloroplast-associated OGs are enriched at the LCA of oomycetes (supplementary additional file 9, Supplementary Material online).
Notably, OGs related to signal transduction, defense and also transcription predominantly expand early in evolution. It has been previously noted that in prokaryotes, the major changes in regulation of transcription and signal transduction often occur at the origins of major lineages (Cordero and Hogeweg 2007). Our observations suggest a similar expansion within the Stramenopile lineage, which may hold true for other eukaryotes as well.
Moreover, OGs characterized as metabolism-related are enriched for duplications at all internal branches throughout the Stramenopiles. Interestingly, OGs related to carbohydrate as well as amino acid transport and metabolism significantly expand at the LCA of oomycetes or throughout the clade. Glycoside hydrolases belong to the class of CAZymes (carbohydrate-active enzymes), which contains proteins involved in synthesis and breakdown of carbohydrates that are found, for example, in the cell wall of both pathogen and host. It has been shown before that glycoside hydrolases are abundant in oomycetes and that the majority of those are potentially secreted (>50%) (Tyler et al. 2006; Ospina-Giraldo et al. 2010; Seidl et al. 2011); however, the evolutionary history of expansion has so far not been uncovered.
Evolutionary Dynamics of Glycoside Hydrolases
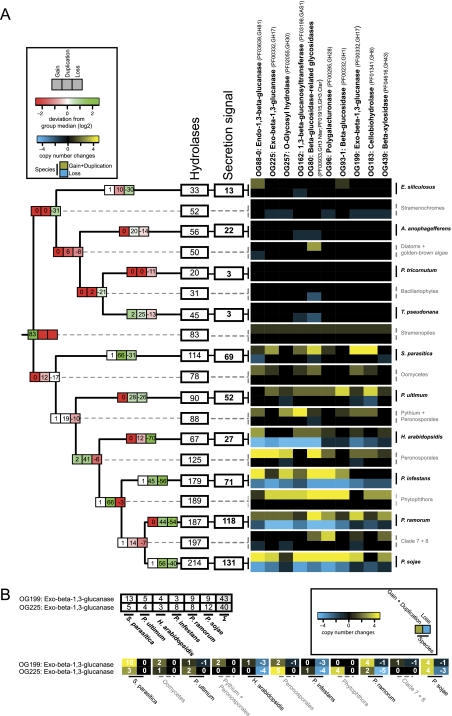
Our systematic analysis of the evolution of glycoside hydrolases revealed that individual OGs that are highly abundant in plant pathogenic oomycetes exhibit distinct evolutionary trajectories (fig. 5A and supplementary additional file 10, Supplementary Material online). Ninety-four OGs are predicted to contain glycoside hydrolases; they cover a total of 1,005 proteins of which the majority (85%) is present in oomycetes (e.g., 179 in P. infestans and 214 in P. sojae) (fig. 4). The repertoire of glycoside hydrolases in oomycetes is dominated by a few large OGs such as, for example, exo-beta-1,3-glucanase (glycoside hydrolase family 17, GH17) (fig. 5A); ∼60% of all glycoside hydrolases in oomycetes belong to only ten OGs. The high abundance of proteins within OGs is not due to isolated duplication events on single branches but is instead caused by consecutive duplications along the internal branches of the oomycete phylogeny. In addition to the high abundance of lineage-specific duplications that are partially balanced by losses, we observed a pronounced accumulation of duplications at the LCA of Peronosporales and especially at the LCA of Phytophthora (66 duplication events).
FIG. 5.—
Global and local pattern of expansion and contraction of OGs containing glycoside hydrolase. (A) The reconciled evolutionary events are projected on the species phylogeny as well as the total abundance of hydrolases at each taxon (ancestral and extant). Heat maps on the different branches display the deviation from the median number of events (i.e., gains, duplications, or losses). The expansion and contraction pattern of the ten largest OGs is displayed next to the phylogeny by a heat map (expansion: yellow; contraction: blue; abundance of duplications/losses saturating at −4 and 4). (B) The number of proteins of two glycoside hydrolase families (OG199 and OG225) in individual species is shown in the table. A heat map displays the expansion and contraction pattern of these two families throughout oomycetes (expansion: yellow; contraction: blue; abundance of duplications/losses saturating at −4 and 4).
Extracellular hydrolases like the exo-beta-1,3-glucanase OG199 and OG225 are examples of OGs that are expanded in oomycetes and lost in Stramenochromes (fig. 5B). The expansion in P. sojae and P. ramorum within OG199 is mainly caused by lineage-specific expansion as well as early duplications followed by subsequent losses in H. arabidopsidis and P. infestans. In contrast, the expansion of OG225 is dominated by consecutive duplications that occur late in evolution, mainly at the LCA of Peronosporales, the LCA of Phytophthora, and lineage specific within P. sojae. These duplications are balanced by subsequent losses in all extant Peronosporales. Even though these OGs share similar biological functions, their high abundance, especially in the Phytophthora spp., is caused by different evolutionary trajectories (fig. 5B).
These OGs do not only differ in their individual evolutionary trajectories but also the whole repertoire of glycoside hydrolases displays a different global pattern of expansion and contraction compared with other functional classes. Another class of highly abundant enzymes in pathogenic oomycetes that have a potential role during infection are peptidases (Tyler et al. 2006; Haas et al. 2009). Whereas the LSCA contains only few glycoside hydrolases (33% of the repertoire observed in P. sojae), many peptidases are already present at the LSCA (225 OGs), and the repertoire of the extant taxa is either of similar size or reduced (supplementary additional file 11, Supplementary Material online). Nevertheless, these peptidase OGs are not static but in constant flux. We demonstrated that pathogenicity related functional classes evolve along different, even opposing trajectories, while still resulting in the observed high abundance in the present-day pathogenic oomycetes.
Discussion
What are the evolutionary events that caused the expansion of OGs in pathogenic oomycetes, and when and how did the dynamic processes that shaped the genome content of these species take place? To address these questions, we systematically studied evolutionary events directly inferred from phylogenetic analysis and tree reconciliation.
Initial work on gene family evolution in Stramenopiles and in particular in pathogenic oomycetes has been limited to a few species and was based on parsimony methods to reconstruct gain and losses of gene families (Martens et al. 2008; Cock et al. 2010). The expansion of families was inferred based on differences in the presence/absence and abundance pattern between species (Tyler et al. 2006; Martens et al. 2008; Haas et al. 2009; Baxter et al. 2010; Lévesque et al. 2010; Seidl et al. 2011). These analyses already provided initial insights into the genome evolution and led to the identification of large gene families that are implied to play a role in host–pathogen interaction. However, the evolutionary trajectories, that is, the patterns of gene gain, duplications, and losses that caused this abundance were not yet systematically analyzed. This study is an additional step toward uncovering these dynamics by a comprehensive phylogenetic analysis and subsequent tree reconciliation of ten Stramenopiles including six pathogenic oomycetes revealing the patterns of gene gains, duplications, and losses that caused this large gene families.
We reconciled the phylome constituted by 18,459 individual protein trees sampled from ten Stramenopiles with a species phylogeny derived by concatenating 189 single-copy genes (fig. 1). The species phylogeny is highly supported and mainly resembles the known topology of the tree of life. It should be noted that the exact topology of the three Phytophthora spp. contradicts the topology published by Blair et al. (2008) that suggested a close association of P. sojae with P. infestans. However, these authors also tested alternatives and concluded that they could not significantly reject the topology in which P. sojae and P. ramorum are closely associated, a grouping that we predict in this study with high support. The number of evolutionary events derived by reconciliation with the topology proposed by Blair et al. (2008) is higher (2,900 events), and hence, our topology is more parsimonious (supplementary additional file 4A, Supplementary Material online). In most cases, reconciliation with either topology did not result in major differences, whereas in some cases, the numbers of evolutionary events are even more pronounced with the topology proposed by Blair et al. (2008), for example, in the case of the accumulation of duplications at the LCA of Phytophthora spp. Recently, Runge et al. (2011) proposed a topology that places H. arabidopsidis as a sister taxon to P. infestans. It has been previously indicated that some clades of Phytophthora are paraphyletic with respect to the downy mildews (Cooke et al. 2000; Göker et al. 2007); however, our reconstructed species phylogeny groups all three analyzed Phytophthora spp. in a single cluster. The number of evolutionary events derived by tree reconciliation with the topology proposed by Runge et al. (2011) is much (∼7,200 events) higher than our more parsimonious topology (supplementary additional file 12, Supplementary Material online). The disagreement between our topology and the two alternatives does not mean that these alternatives are wrong. Nevertheless, we preferred to use the phylogeny that was reconstructed from our concatenated alignment containing 189 loci. When reconciling a large number of gene families, this topology is the most parsimonious and hence conservative, therefore further supporting our choice.
A comprehensive and dynamic picture of the genome evolution in Stramenopiles was obtained by projecting gene gains, duplications, and losses that were derived by reconciliation of the phylome onto the species phylogeny (fig. 2). Our analysis demonstrates that throughout evolution, the genomes of Stramenopiles are not static but in constant flux; a dynamic that is at least partially disguised by parsimonious-based methods when duplications and losses occurred in the same OG at the same branch. Whereas the genome content of Stramenochromes is of comparable size to the LSCA, genomes of pathogenic oomycetes have been growing by gains and by continuous duplications on both the internal as well as the terminal branches. The LSCA is large and contained ∼10,000 genes of which the majority predate the LSCA.
Some of these genes might have not transferred vertically but instead descend from a horizontal gene transfer (HGT). Consequently, we may overestimate the number of genes in the LSCA, introduce unnecessary losses in the derived lineages, and underestimate gains in internal branches. So far, there are only few comprehensive studies that have investigated the fraction of HGTs in Stramenopiles from origins, such as bacteria or eukaryotes (Richards et al. 2006, 2011; Richards and Talbot 2007; Morris et al. 2009). A recent analysis of HGT between fungi and oomycetes has revealed 34 high-confidence HGTs that together contributed to up to ∼8% of the secretome of P. ramorum and hence to plant parasitic mechanisms of oomycetes (Richards et al. 2011). Indeed, one of their discussed examples, a sugar transporter called AraJ (Richards et al. 2006, 2011), is annotated as ancestral (gained at the LSCA or before) in our analysis. More quantitatively, if we consider all OGs that consistently have their best blast hits to eukaryotes or bacteria as potential sources of HGT, only a minority (excluding singletons because these are not considered in our reconstruction) is specific to either oomycetes or Stramenochromes (supplementary additional file 13, Supplementary Material online). These are the only cases where an erroneous placement of the gains at the LSCA could influence our results because OGs that have members in both lineages will be invariably placed at the LCSA. These numbers are of course upper limits because real losses of ancestral OGs at either ancestor of the two lineages also occur or are included in the reported numbers (supplementary additional file 13, Supplementary Material online). Consequently, the quantitative influence of these events to our analysis is marginal, even though it highlights the mosaic nature of the analyzed species.
The interpretation of the inferred gene content of LSCA and the genome evolution of Stramenopiles also depends on the contribution of the plastid to their gene content. If the LCA contained a plastid, as proposed by the Chromalveolate hypothesis (Cavalier-Smith 1999; Keeling 2009), then our estimated size of the LSCA as well as the derived evolutionary events do not change (fig. 2). However, our results would be affected if the acquisition of the plastid by the photosynthetic Stramenochromes occurred after the speciation of oomycetes as suggested by the SEEE hypothesis (Cavalier-Smith et al. 1994; Archibald 2009; Baurain et al. 2010). If the plastid endosymbiosis mainly affected chloroplast-associated genes, we would slightly overestimate the size of the LSCA by 295 genes (2.8%) and an equivalent number of losses and gains at the branches leading to oomycetes and Stramenochromes (supplementary additional file 8, Supplementary Material online). However, if the plastid endosymbiosis contributed a wide array of cellular functions to the Stramenochrome ancestor, we would overestimate the size of the LSCA by up to 2,300 genes (fig. 2). This number has to be seen as the upper limit because we obtained it by assuming that every OG that we inferred to be lost at the branch leading to oomycetes has descended from the plastid endosymbiosis (fig. 2). In contradiction to the SEEE hypothesis, we observed 432 OGs that are chloroplast-associated and retained in the genomes of both nonphotosynthetic oomycetes and Stramenochromes since the LSCA (supplementary additional file 8, Supplementary Material online). Similarly, 88 and 14 oomycete-specific OGs have their best blast hits in green and red algae genomes, respectively (supplementary additional file 13, Supplementary Material online). These results, together with studies by others (Andersson and Roger 2002; Tyler et al. 2006; Maruyama et al. 2009), seem to slightly favor the early acquisition of the plastid before the speciation of Stramenochromes and oomycetes. However, recent molecular data support a more complex scenario and later acquisition of the plastid thereby rejecting the Chromalveolate hypothesis (Stiller et al. 2009; Baurain et al. 2010; Felsner et al. 2011; Woehle et al. 2011). Nevertheless, our results do not change dramatically and are hence independent of the precise history of the plastid. Dedicated future research, also facilitated by additional genomes from related lineages, will gather additional evidence for either of the two hypotheses and thereby shed light on this controversially discussed event and hence also on our reconstructions.
The massive accumulation of duplications at the LCA of Phytophthora spp. points to a large-scale duplication event (fig. 3; supplementary additional file 6, Supplementary Material online). It has been postulated that the accumulation of duplications at a constrained point in time can be indicative for duplications that affect either large parts of the genome or the whole genome (McLysaght et al. 2002; Jaillon et al. 2004; Kellis et al. 2004; Jiao et al. 2011). This accumulation of duplication events was already observed earlier by Martens and colleagues who used an independent method to time the age of paralogs in Phytophthora spp. (Martens and Van de Peer 2010). The usage of additional outgroup species allows us to more precisely estimate the time of these events, which seem to have happened after the speciation of H. arabidopsidis and before the radiation of Phytophthora spp. Nevertheless, the usage of the less parsimonious topology of the analyzed Peronosporales proposed by Runge et al. (2011) introduces an accumulation of duplications at the LCA of Peronosporales (supplementary additional file 12, Supplementary Material online). Hence, if this proposed topology is correct, it is tempting to speculate that the analyzed Peronosporales shared this large-scale duplication event. Considering our predicted topology, such an earlier timing of this event could also be possible; the genome contraction of H. arabidopsidis might lead to the loss of both duplicates and hence at least partially obscure events happening at the LCA of Peronosporales. Nevertheless, neither the analysis performed by Martens and colleagues nor ours is able to elucidate the exact mode of expansion because of the lack of long-distance intra-species collinearity of genes. Alternative scenarios, such as segmental duplications that occurred at a constrained point in time followed by reorganization, are at least equally likely, especially given the observed dynamics in genome organization of Phytophthora spp. and the genome contraction in H. arabidopsidis. Independent of the underlying mechanism, this coordinated expansion of gene families marks a major transition point in their evolution. Together with subsequent lineage-specific losses, the expansion could be the driving force of the speciation and adaptation to different hosts within the Phytophthora genus (or even within the Peronosporales); a process that has been proposed before for other organisms, such as yeast (Kellis et al. 2004) or plants (Jiao et al. 2011).
The number of duplications and losses events at each branch is determined by tree reconciliation. This procedure is not only dependent on a reliable species phylogeny, but also on the alignment as well as the gene tree, or in this case, protein tree. Erroneously inferred protein trees, either based on inaccurate alignments or due to biases in the tree predictions itself, will artificially increase the number of duplications at internal branches and losses at terminal branches of the tree. To address if the incorporation of low-quality alignments in our analysis interferes with our main results, we divided the families into high-quality and low-quality alignments (see supplementary additional files 2 and 16A, Supplementary Material online). If we remove the 477 families and their derived OGs that have a low-quality alignment in our analysis, we observe that the absolute numbers of evolutionary events decrease as the analysis is now based on less data (supplementary additional file 16B, Supplementary Material online). More importantly, the relative numbers and the major trends observed in our analysis, such as the accumulation of duplication in the common ancestor of Phytophthora spp., are independent of the exclusion of the lower quality alignments (supplementary additional file 16B, Supplementary Material online). Consequently, our results are robust to the possible bias introduced by the retention of the full set of families. To reduce the possible bias in the tree prediction and to apply an explicit model of evolution, we used a maximum likelihood method to predict the tree topology of the protein trees. More importantly, we used NOTUNG (Chen et al. 2000; Durand et al. 2006) for tree reconciliation that allows to explicitly address this uncertainty in protein trees. NOTUNG allows the rearrangement of weakly supported parts of the tree topology to reduce the evolutionary events needed for reconciliation while keeping strongly supported parts fixed. Throughout this study, we used a bootstrap support of >80% to indicate strongly supported clades of the protein trees. Hence, parts of the tree topology that are not supported with a bootstrap of at least 80% are rearranged to minimize evolutionary events. When we compared the results derived with >80% cutoff to a less conservative cutoff of >60%, leading to less rearrangement, we indeed observed more duplications at the internal branches and more losses at terminal branches, especially within oomycetes (supplementary additional file 14A, Supplementary Material online). When we applied an even stricter cutoff of >90%, which resulted in more rearrangement, some duplications at the internal branches, for example, at the LCA of Stramenochromes and especially at the LCA of Peronosporales, were removed; consequently, fewer losses in the terminal taxa were introduced (supplementary additional file 14B, Supplementary Material online). Regardless of the choice of the cutoff (60%, 80%, or 90%), the changes in the abundance of the reconciled evolutionary events did not interfere with our global results indicating the robustness of our framework to this bias.
Our results are directly dependent on the availability, quality, and completeness of the predicted proteomes derived from the various sequenced genomes. The robustness of gene annotation has been observed to have only small effects on the analysis of gene family losses in related species (Martens et al. 2008). In general, more sequenced genomes of closely related oomycetes, preferably sister taxa to the already existing genomes, would enable a more precise timing of the duplication events, especially at the terminal branches. Moreover, our analyses are currently limited to pathogenic oomycetes. Including sequenced genomes of saprophytic species would elucidate whether evolutionary events at the LCA of oomycetes are specific to pathogenic oomycetes or are instead a general pattern for all oomycetes.
Conclusions
We systematically analyzed the genome evolution of pathogenic oomycetes by reconciliation of the Stramenopile phylome with a highly supported species phylogeny. Our analysis uncovered that oomycete genomes, emanating from a common ancestor of Stramenopiles that had a rather large genome encoding for ∼10,000 genes, were growing by continuous duplications that predominantly affected ancestral OGs. The massive accumulation of duplication events at the LCA of the Phytophthora genus suggests a large-scale duplication event that predates the speciation and hence might be driving the adaptive radiation within this genus. Different functional classes have distinct evolutionary trajectories: not only between classes but also within a single class. Different evolutionary trajectories are proposed to lead to the observed abundance of pathogenicity-related functional classes, for example, glycoside hydrolases and peptidases, an observation that was not yet apparent by previous analyses. Consequently, we unveiled both large-scale evolutionary processes that shape the genomes of extant oomycetes as well as the complex evolution trajectories that lead to highly abundant gene families in this important class of pathogens.
Supplementary Material
Supplementary additional files 1–16 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank Lidija Berke, Like Fokkens, Adrian Schneider, and Gabino F. Sanchez-Perez for helpful discussions and comments on the manuscript and Stefan Zoller for technical support. Some of the sequence data and annotation were provided by the US Department of Energy Joint Genome Institute (http://www.jgi.doe.gov), the Broad Institute of Harvard, and the Massachusetts Institute of Technology (http://www.broadinstitute.org) in collaboration with the user community (for detailed information, see supplementary additional file 2, Supplementary Material online). This project was financed by the Centre for BioSystems Genomics (CBSG) which is part of the Netherlands Genomics Initiative/Netherlands Organisation for Scientific Research.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Andersson JO, Roger AJ. A cyanobacterial gene in nonphotosynthetic protists—an early chloroplast acquisition in eukaryotes? Curr Biol. 2002;12:115–119. doi: 10.1016/s0960-9822(01)00649-2. [DOI] [PubMed] [Google Scholar]
- Archibald JM. The puzzle of plastid evolution. Curr Biol. 2009;19:R81–R88. doi: 10.1016/j.cub.2008.11.067. [DOI] [PubMed] [Google Scholar]
- Baurain D, et al. Phylogenomic evidence for separate acquisition of plastids in cryptophytes, haptophytes, and stramenopiles. Mol Biol Evol. 2010;27:1698–1709. doi: 10.1093/molbev/msq059. [DOI] [PubMed] [Google Scholar]
- Baxter L, et al. Signatures of adaptation to obligate biotrophy in the Hyaloperonospora arabidopsidis genome. Science. 2010;330:1549–1551. doi: 10.1126/science.1195203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair JE, Coffey MD, Park S-Y, Geiser DM, Kang S. A multi-locus phylogeny for Phytophthora utilizing markers derived from complete genome sequences. Fungal Genet Biol. 2008;45:266–277. doi: 10.1016/j.fgb.2007.10.010. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Principles of protein and lipid targeting in secondary symbiogenesis: euglenoid, dinoflagellate, and sporozoan plastid origins and the eukaryote family tree. J Eukaryot Microbiol. 1999;46:347–366. doi: 10.1111/j.1550-7408.1999.tb04614.x. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T, Allsopp MT, Chao EE. Chimeric conundra: are nucleomorphs and chromists monophyletic or polyphyletic? Proc Natl Acad Sci U S A. 1994;91:11368–11372. doi: 10.1073/pnas.91.24.11368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Durand D, Farach-Colton M. NOTUNG: a program for dating gene duplications and optimizing gene family trees. J Comput Biol. 2000;7:429–447. doi: 10.1089/106652700750050871. [DOI] [PubMed] [Google Scholar]
- Cock JM, et al. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature. 2010;465:617–621. doi: 10.1038/nature09016. [DOI] [PubMed] [Google Scholar]
- Conesa A, et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Cooke D, Drenth A, Duncan J, Wagels G, Brasier C. A Molecular Phylogeny of Phytophthora and Related Oomycetes. Fungal Genet Biol. 2000;30:17–32. doi: 10.1006/fgbi.2000.1202. [DOI] [PubMed] [Google Scholar]
- Cordero OX, Hogeweg P. Large changes in regulome size herald the main prokaryotic lineages. Trends Genet. 2007;23:488–493. doi: 10.1016/j.tig.2007.07.006. [DOI] [PubMed] [Google Scholar]
- Dou D, et al. RXLR-mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen-encoded machinery. Plant Cell. 2008;20:1930–1947. doi: 10.1105/tpc.107.056093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duplessis S, et al. Obligate biotrophy features unraveled by the genomic analysis of rust fungi. Proc Natl Acad Sci U S A. 2011;108:9166–9171. doi: 10.1073/pnas.1019315108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand D, Halldórsson BV, Vernot B. A hybrid micro-macroevolutionary approach to gene tree reconstruction. J Comput Biol. 2006;13:320–335. doi: 10.1089/cmb.2006.13.320. [DOI] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsner G, et al. ERAD components in organisms with complex red plastids suggest recruitment of a preexisting protein transport pathway for the periplastid membrane. Genome Biol Evol. 2011;3:140–150. doi: 10.1093/gbe/evq074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch WM. Homology a personal view on some of the problems. Trends Genet. 2000;16:227–231. doi: 10.1016/s0168-9525(00)02005-9. [DOI] [PubMed] [Google Scholar]
- Gijzen M, Nürnberger T. Nep1-like proteins from plant pathogens: recruitment and diversification of the NPP1 domain across taxa. Phytochemistry. 2006;67:1800–1807. doi: 10.1016/j.phytochem.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Gobler CJ, et al. Niche of harmful alga Aureococcus anophagefferens revealed through ecogenomics. Proc Natl Acad Sci U S A. 2011;108:4352–4357. doi: 10.1073/pnas.1016106108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göker M, Voglmayr H, Riethmüller A, Oberwinkler F. How do obligate parasites evolve? A multi-gene phylogenetic analysis of downy mildews. Fungal Genet Biol. 2007;44:105–122. doi: 10.1016/j.fgb.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Govers F, Bouwmeester K. Effector trafficking: RXLR-dEER as extra gear for delivery into plant cells. Plant Cell. 2008;20:1728–1730. doi: 10.1105/tpc.108.062075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govers F, Gijzen M. Phytophthora genomics: the plant destroyers' genome decoded. Mol Plant Microbe Interact. 2006;19:1295–1301. doi: 10.1094/MPMI-19-1295. [DOI] [PubMed] [Google Scholar]
- Haas BJ, et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 2009;461:393–398. doi: 10.1038/nature08358. [DOI] [PubMed] [Google Scholar]
- Jaillon O, et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- Jiang RHY, Tripathy S, Govers F, Tyler BM. RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proc Natl Acad Sci U S A. 2008;105:4874–4879. doi: 10.1073/pnas.0709303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang RHY, et al. Elicitin genes in Phytophthora infestans are clustered and interspersed with various transposon-like elements. Mol Genet Genomics. 2005;273:20–32. doi: 10.1007/s00438-005-1114-0. [DOI] [PubMed] [Google Scholar]
- Jiao Y, et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- Kale SD, et al. External lipid PI3P mediates entry of eukaryotic pathogen effectors into plant and animal host cells. Cell. 2010;142:284–295. doi: 10.1016/j.cell.2010.06.008. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K-I, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ. Chromalveolates and the evolution of plastids by secondary endosymbiosis. J Eukaryot Microbiol. 2009;56:1–8. doi: 10.1111/j.1550-7408.2008.00371.x. [DOI] [PubMed] [Google Scholar]
- Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- Latijnhouwers M, de Wit PJGM, Govers F. Oomycetes and fungi: similar weaponry to attack plants. Trends Microbiol. 2003;11:462–469. doi: 10.1016/j.tim.2003.08.002. [DOI] [PubMed] [Google Scholar]
- Lévesque CA, et al. Genome sequence of the necrotrophic plant pathogen Pythium ultimum reveals original pathogenicity mechanisms and effector repertoire. Genome Biol. 2010;11:R73. doi: 10.1186/gb-2010-11-7-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens C, Van de Peer Y. The hidden duplication past of the plant pathogen Phytophthora and its consequences for infection. BMC Genomics. 2010;11:353. doi: 10.1186/1471-2164-11-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens C, Vandepoele K, Van de Peer Y. Whole-genome analysis reveals molecular innovations and evolutionary transitions in chromalveolate species. Proc Natl Acad Sci U S A. 2008;105:3427–3432. doi: 10.1073/pnas.0712248105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama S, Matsuzaki M, Misawa K, Nozaki H. Cyanobacterial contribution to the genomes of the plastid-lacking protists. BMC Evol Biol. 2009;9:197. doi: 10.1186/1471-2148-9-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLysaght A, Hokamp K, Wolfe KH. Extensive genomic duplication during early chordate evolution. Nat Genet. 2002;31:200–204. doi: 10.1038/ng884. [DOI] [PubMed] [Google Scholar]
- Morris PF, et al. Multiple horizontal gene transfer events and domain fusions have created novel regulatory and metabolic networks in the oomycete genome. PLoS One. 2009;4:e6133. doi: 10.1371/journal.pone.0006133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, et al. eggNOG v2.0: extending the evolutionary genealogy of genes with enhanced non-supervised orthologous groups, species and functional annotations. Nucleic Acids Res. 2010;38:D190–D195. doi: 10.1093/nar/gkp951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ospina-Giraldo MD, Griffith JG, Laird EW, Mingora C. The CAZyome of Phytophthora spp.: a comprehensive analysis of the gene complement coding for carbohydrate-active enzymes in species of the genus Phytophthora. BMC Genomics. 2010;11:525. doi: 10.1186/1471-2164-11-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson D. The diversity of eukaryotes. Am Nat. 1999;154:96–124. doi: 10.1086/303287. [DOI] [PubMed] [Google Scholar]
- Richards TA, Dacks JB, Jenkinson JM, Thornton CR, Talbot NJ. Evolution of filamentous plant pathogens: gene exchange across eukaryotic kingdoms. Curr Biol. 2006;16:1857–1864. doi: 10.1016/j.cub.2006.07.052. [DOI] [PubMed] [Google Scholar]
- Richards TA, Talbot NJ. Plant parasitic oomycetes such as phytophthora species contain genes derived from three eukaryotic lineages. Plant Signal Behav. 2007;2:112–114. doi: 10.4161/psb.2.2.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards TA, et al. Horizontal gene transfer facilitated the evolution of plant parasitic mechanisms in the oomycetes. Proc Natl Acad Sci U S A. 2011;108(37):15258–15263. doi: 10.1073/pnas.1105100108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runge F, et al. The inclusion of downy mildews in a multi-locus-dataset and its reanalysis reveals a high degree of paraphyly in Phytophthora. IMA Fungus. 2011;2:163–171. doi: 10.5598/imafungus.2011.02.02.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidl MF, Van den Ackerveken G, Govers F, Snel B. A domain-centric analysis of oomycete plant pathogen genomes reveals unique protein organization. Plant Physiol. 2011;155:628–644. doi: 10.1104/pp.110.167841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer ELL, Koonin EV. Orthology, paralogy and proposed classification for paralog subtypes. Trends Genet. 2002;18:619–620. doi: 10.1016/s0168-9525(02)02793-2. [DOI] [PubMed] [Google Scholar]
- Spanu PD, et al. Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science. 2010;330:1543–1546. doi: 10.1126/science.1194573. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stassen JH, Van den Ackerveken G. How do oomycete effectors interfere with plant life? Curr Opin Plant Biol. 2011;14:1–8. doi: 10.1016/j.pbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Stiller JW, Huang J, Ding Q, Tian J, Goodwillie C. Are algal genes in nonphotosynthetic protists evidence of historical plastid endosymbioses? BMC Genomics. 2009;10:484. doi: 10.1186/1471-2164-10-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Tyler BM, et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006;313:1261–1266. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- Van Dongen S. A cluster algorithm for graphs. 2000. Report INS-R0010. Amsterdam: National Research Institute for Mathematics and Computer Science in the Netherlands. [Google Scholar]
- Whisson SC, et al. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature. 2007;450:115–118. doi: 10.1038/nature06203. [DOI] [PubMed] [Google Scholar]
- Woehle C, Dagan T, Martin WF, Gould SB. Red and problematic green phylogenetic signals among thousands of nuclear genes from the photosynthetic and apicomplexa-related Chromera velia. Genome Biol Evol. 2011;3:1220–1230. doi: 10.1093/gbe/evr100. [DOI] [PMC free article] [PubMed] [Google Scholar]