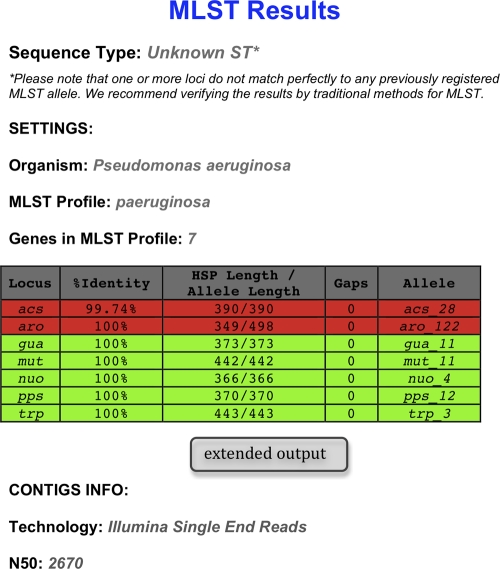
Fig 1.
MLST results for a P. aeruginosa isolate in the short output format. By use of the MLST Web server, a P. aeruginosa strain that had been sequenced on the Illumina platform generating single reads was typed. For the purpose of the example, we have chosen to show the results obtained by using short sequence reads that assemble into a draft genome with a low N50. Shown are the name of the loci in the MLST scheme, the percentage of nucleotides that are identical in the best-matching MLST allele in the database and the corresponding sequence in the genome (% identity), the length of the alignment between the best-matching MLST allele in the database and the corresponding sequence in the genome (also called the high-scoring segment pair [HSP]), the length of the best-matching MLST allele in the database, the number of gaps in the HSP, and the name of the best-matching MSLT allele. Note that for a perfectly matching allele, the percentage of identity will be 100%, the allele length will equal the HSP length, and the number of gaps will be zero. Green indicates a perfect match, while red indicates an imperfect match.