Fig 2.
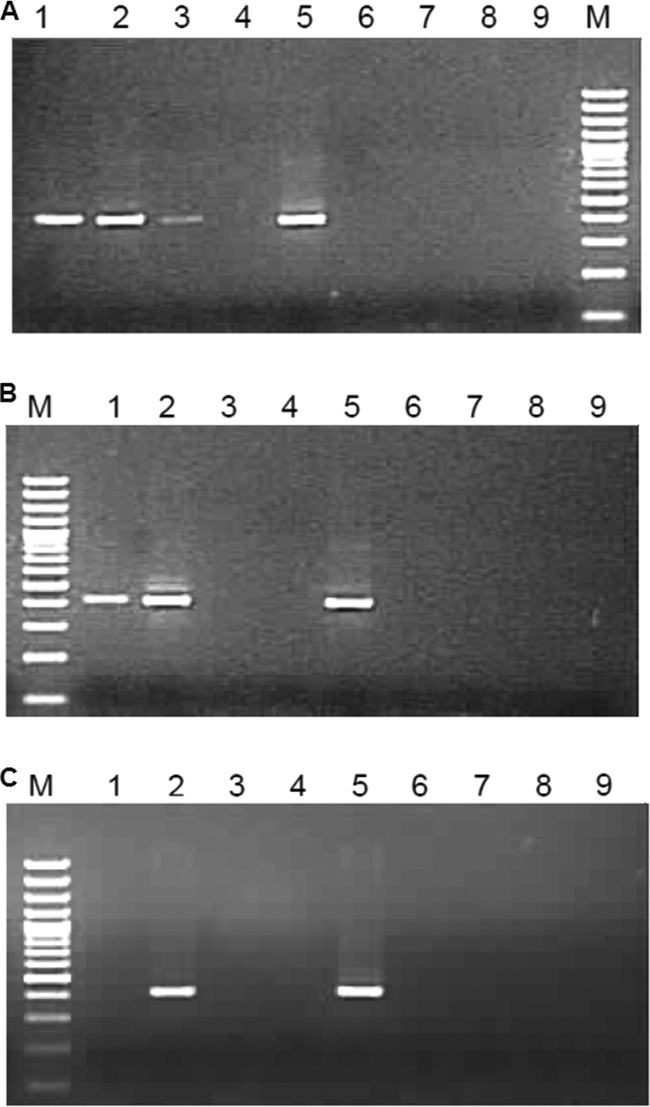
Tag-based PCR to remove false priming: strand-specific assays were performed on 106 (A), 105 (B), and 103 (C) full-genome positive-sense RNA copies by the use of two reverse transcriptase enzymes. For each gel, lanes 1 to 3 represent reactions with AMV-RT (lane 1, RT with HEVTagF1 primer; lane 2, RT with HEVTagR1 primer; lane 3, no primer), lanes 4 to 6 represent reactions with Superscript RT-III (lane 4, RT with HEVTagF1 primer; lane 5, RT with HEVTagR1 primer; lane 6, no primer), and lanes 7 to 9 represent reactions without any RT enzyme (lane 7, HEVTagF1 primer; lane 8, HEVTagR1 primer; lane 9, no primer). After reverse transcription and exonuclease I treatment, cDNAs were purified by the use of a silica-based column. Samples without reverse transcriptase enzyme were processed similarly to samples with RT enzymes.
