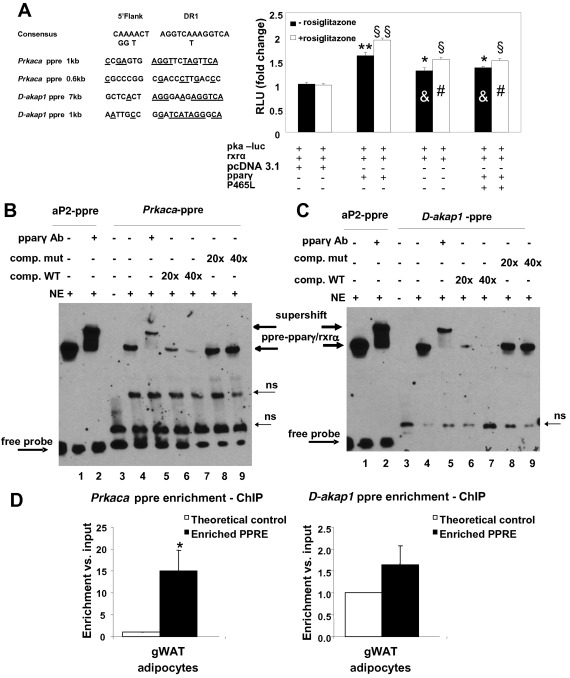
Fig 2.
Transactivation of the PKAcat subunit α promoter-luciferase reporter construct (pka-luc) by PPARγ and the PPARγ P465L mutant in HEK293T cells. (A) PPARγ transcriptionally activates prkaca. The pka-luc reporter plasmid was transiently cotransfected with a plasmid expressing RXRα together with PPARγ, the P465L mutant, or a combination of both into HEK293T cells. At 24 h posttransfection, the cells were incubated with 1 μM rosiglitazone (open bars) or vehicle (solid bars) and further incubated for 24 h before being harvested and assayed for luciferase activity. Here, we show a representative experiment out of 3 independent experiments (n = 12 for each experimental condition). The luciferase activity measured in cells transfected with pka-luc and RXRα with pcDNA 3.1 as a control for PPARγ and the P465L mutant (basal condition) in the absence of rosiglitazone was set at 1. **, P < 0.0001, and *, P < 0.001 compared to the basal condition. &, P < 0.001 compared to PPARγ transfection (− rosiglitazone). §§, P < 0.0001, and §, P < 0.001 compared to the basal condition (+ rosiglitazone). #, P < 0.0001 compared to PPARγ transfection (+ rosiglitazone). (B and C) PPARγ binds a new PPRE identified in prkaca and d-akap1 promoters. Shown are electrophoretic mobility shift and supershift assays on putative PPRE of the murine PKA (mPKA) catalytic subunit α promoter and the mAKAP1 promoter. The 3 biotin-labeled double-stranded oligonucleotide probes corresponding to the mouse PKAcat subunit α PPRE (prkaca-ppre) (B) and d-akap-PPRE (C) were incubated with 5 μg of nuclear extract (NE) of HEK293T cells overexpressing PPARγ and RXRα in the absence or presence of PPAR antibody. The gel was transferred to a nylon membrane, and the shifted bands were detected by incubating the membrane with streptavidin-horseradish peroxidase, followed by chemiluminescence detection. Lanes 1, biotin-labeled double-stranded aP2-ppre probe incubated with nuclear extract; lanes 2, biotin-labeled aP2-ppre probe incubated with nuclear extract in the presence of PPAR antibody; lanes 3, PKAcat α-ppre or D-AKAP1 biotinylated probes alone; lanes 4, PKAcat α-ppre or d-akap1 biotinated probes with nuclear extract; lane 5, PKAcat α-ppre or d-akap1 biotinated probes incubated with nuclear extract in the presence of PPAR antibody; lanes 6 and 7, for both PKAcatα and D-AKAP1, 20× and 40× excesses of unlabeled double-stranded wt oligonucleotides were included as competitors with nuclear extract and labeled wt probe; lanes 8 and 9, for both PKAcatα and D-AKAP1, 20× and 40× excesses of unlabeled double-stranded mutated oligonucleotides were included as competitors with nuclear extract and labeled wt probes. (D) PPARγ binds a new PPRE identified in the prkaca promoter in vivo. Shown is a chromatin immunoprecipitation assay of the putative PPRE of the mPKAcat subunit α promoter and mAKAP1 promoter. Isolated mature adipocytes from gonadal pads of wt mice were collected, and ChIP assays were performed, using an anti-PPARγ antibody to immunoprecipitate PPARγ-linked DNA. Quantification of PPRE sequences for both prkaca and d-akap1 promoters was performed by real-time PCR, considering the fold change with respect to a control sequence in the same gene and normalized to the input DNA as described in Materials and Methods. The data are expressed as means and standard errors of the mean (SEM) from six independent experiments. gWAT, gonadal WAT.