Abstract
We report that Rcf1 (formerly Aim31), a member of the conserved hypoxia-induced gene 1 (Hig1) protein family, represents a novel component of the yeast cytochrome bc1-cytochrome c oxidase (COX) supercomplex. Rcf1 (respiratory supercomplex factor 1) partitions with the COX complex, and evidence that it may act as a bridge to the cytochrome bc1 complex is presented. Rcf1 interacts with the Cox3 subunit and can do so prior to their assembly into the COX complex. A close proximity of Rcf1 and members of the ADP/ATP carrier (AAC) family was also established. Rcf1 displays overlapping function with another Hig1-related protein, Rcf2 (formerly Aim38), and their joint presence is required for optimal COX enzyme activity and the correct assembly of the cytochrome bc1-COX supercomplex. Rcf1 and Rcf2 can independently associate with the cytochrome bc1-COX supercomplex, indicating that at least two forms of this supercomplex exist within mitochondria. We provide evidence that the association with the cytochrome bc1-COX supercomplex and regulation of the COX complex are a conserved feature of Hig1 family members. Based on our findings, we propose a model where the Hig1 proteins regulate the COX enzyme activity through Cox3 and associated Cox12 protein, in a manner that may be influenced by the neighboring AAC proteins.
INTRODUCTION
Mitochondria are responsible for the aerobic production of ATP through a process termed oxidative phosphorylation (OXPHOS) (29, 35, 39). The enzyme complexes involved in the OXPHOS pathway are embedded in the mitochondrial inner membrane and include the electron transport chain complexes (ETC) and the F1Fo-ATP synthase. Cytochrome c oxidase (COX) is the terminal member of the ETC and passes electrons from cytochrome c to O2, generating water (13, 30, 35, 41). The ATP generated in the mitochondrial matrix is shuttled out of the mitochondria, in exchange for ADP, through the ADP/ATP carrier (AAC) protein family (33). The ETC physically associate with each other in the membrane to form supercomplexes, e.g., the cytochrome bc1-COX supercomplex, which is thought to enhance the electron transfer and regulation of these complexes (8, 12, 20, 29, 35, 39, 40). Evidence for the physical association of the AAC proteins with the cytochrome bc1-COX supercomplex has also been reported, and this may represent a mechanism for communication between the levels of OXPHOS activity and the ATP/ADP levels in the mitochondria (7, 11, 40).
Both the cytochrome bc1 and COX complexes are oligomeric complexes of mosaic origin. In the case of the COX complex, the three central core subunits, Cox1, Cox2, and Cox3, are encoded by the mitochondrial DNA (mtDNA; [rho]) and are homologous to the corresponding COX enzyme subunits in bacteria (9, 41). The remaining COX subunits are encoded in the nucleus and imported into mitochondria, where they are assembled in an organized and multistep fashion with the proteins Cox1, -2, and -3 (9, 13–15, 30, 31). A significant amount of information is known about the assembly and function of the Cox1 and Cox2 proteins, and specific assembly chaperones of these proteins have been reported (13, 30, 41). No assembly partner for Cox3 has been identified to date, in either bacterial or mitochondrial systems.
A number of independent studies in diverse organisms aimed at identifying genes whose expression is induced during hypoxia have led to the discovery of a gene family termed the hypoxia-induced gene 1 (Hig1) family (1, 10, 17, 19, 38, 42). The Hig1 family is conserved throughout evolution, with members in the alphaproteobacteria and diverse eukaryotic taxa, including plants, fungi, nematodes, and mammals. Elevated expression of Hig1 family members was also found in response to other stress inducers, including low glucose and light and oxidative stress, and in hypoxic cervical carcinoma cells (1, 10, 17, 19, 38, 42). Preliminary studies indicate that some Hig1 isoforms may exert an antiapoptotic function, in particular in response to prolonged exposure to oxidative and/or hypoxic stress (1, 38, 42). Although one study showed a Hig1 isoform to be a mitochondrial inner membrane protein (42), the function(s) of these conserved Hig1 proteins is currently unknown.
In the yeast Saccharomyces cerevisiae, the Aim31 protein displays a high degree of similarity to members of the Hig1 protein family. Originally discovered in a screen designed to identify genes encoding proteins whose absence caused an altered inheritance of mtDNA (AIM) (22), the function of Aim31 was unknown. Here we report that Aim31 can be found in association with the cytochrome bc1-COX supercomplex, where it appears to bind to both aspects of this complex, i.e., to both the cytochrome bc1 and COX enzyme domains. We demonstrate that Aim31 is more tightly associated with the COX complex, where it displays a close physical relationship with the Cox3 protein. Furthermore, we show that Aim31 shares an overlapping function with a second mitochondrial protein with limited similarity to Aim31 and Hig1 proteins, the Aim38 protein. Our data indicate that Aim31 and Aim38 may independently bind to the cytochrome bc1-COX supercomplex and that loss of both Aim31 and Aim38 (but not loss of only one of them) has a significant impact on the COX enzyme activity and assembly of the peripheral COX subunits Cox12 and Cox13. The presence of Aim31 and Aim38 proteins is required for the correct assembly of the cytochrome bc1-COX supercomplex, and we propose that they may act as bridges to support the assembly of the supercomplex state. Given their functional relevance for the COX enzyme and their physical association with the cytochrome bc1-COX supercomplex, we propose renaming the Aim31 and Aim38 proteins Rcf1 and Rcf2, respectively (Rcf is for respiratory supercomplex factors). Together, our findings shed light on the important role the conserved Hig1 protein family plays in the regulation and functioning of mitochondrial COX complex, the most abundant oxygen-consuming enzyme of the cell.
MATERIALS AND METHODS
Yeast strains and growth conditions.
The YML030w/AIM31 and the YNR018w/AIM38 genes are referred to here as the RCF1 and RCF2 genes, respectively. S. cerevisiae strains used in this study are the wild type (WT; W303-1A mata leu2 trp1 ura3 his3 ade2), an RCF1::HIS3 (Δrcf1) strain (W303-1A mata leu2 trp1 ura3 ade2), an RCF2::KAN (Δrcf2) strain (W303-1B matα leu2 trp1 ura3 his3 ade2), and an RCF1::HIS3 RCF2::KAN (Δrcf1 Δrcf2) strain (W303-1A mata leu2 trp1 ura3 ade2). The double mutant was created by mating the respective single mutant haploid strains and followed by tetrad dissection of the resulting diploid strain. Yeast strains were maintained and cultured using standard protocols at 30°C on YP medium supplemented with 2% glucose and 20 mg/liter adenine hemisulfate (YPAD). Mitochondria were isolated from cultures grown in YP-0.5% lactate medium supplemented with 2% galactose (YP-Gal).
Expression of the His-tagged protein derivatives.
For the expression of His-tagged proteins, a DNA fragment encoding Rcf1His, Rcf1ΔCHis (residues 1 to 112), Rcf2His, M05D6.5His, Aac1His, and Aac3His (each bearing a 12-histidine tag at the C terminus) was generated through PCR using yeast genomic DNA or Caenorhabditis elegans cDNA as a template and was cloned as an XbaI/PstI fragment into a Yip351-based vector downstream of the galactose-inducible GAL10 promoter. The recombinant plasmids were integrated into the yeast genome of indicated strains at the leu2 gene locus following linearization with BstEII. Expression of the His-tagged proteins was induced by the inclusion of galactose in the growth medium, as indicated in the legend to Fig. 3. and was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), Western blotting, and immunodecoration of mitochondrial extracts from resulting transformants. The creation and expression of the HisAac2 derivative have been previously described (11).
Fig 3.
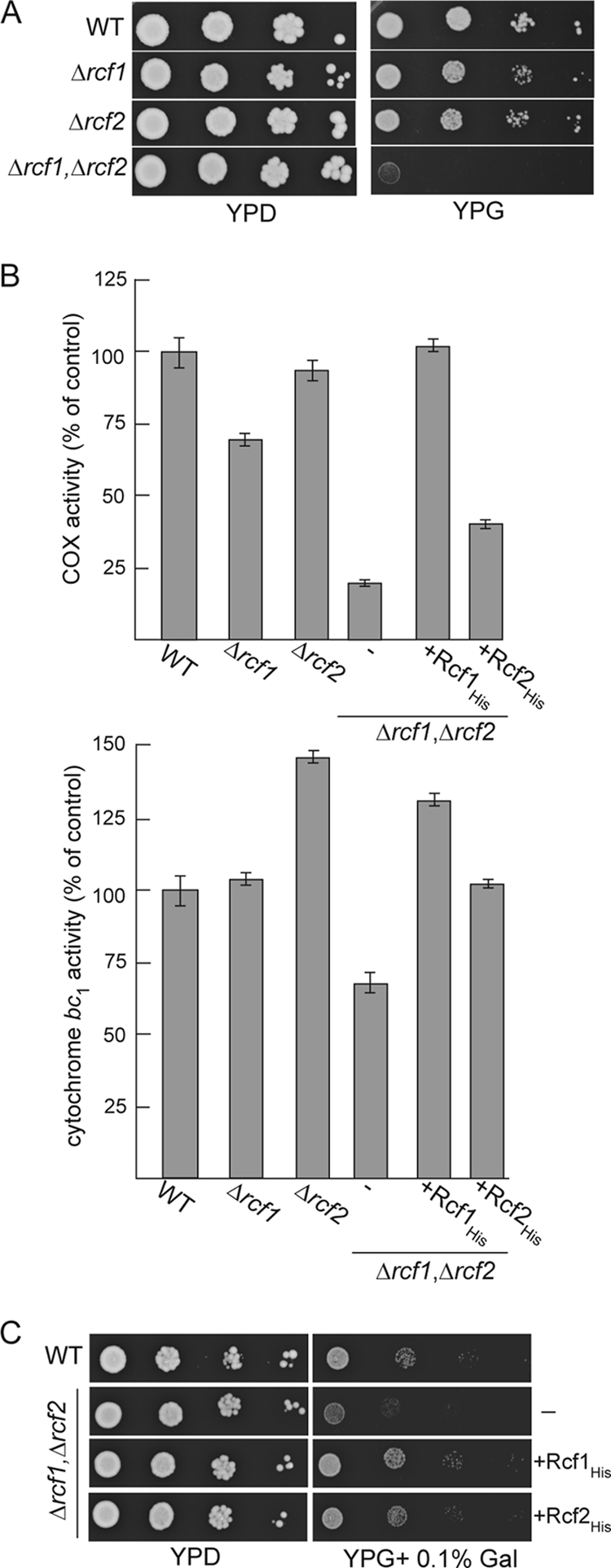
RCF1 and RCF2 genetically interact and together are required to support the COX enzyme activity. (A) Serial 10-fold dilutions of wild-type (WT), Δrcf1, Δrcf2, or Δrcf1 Δrcf2 cells were spotted on YP plates containing glucose (YPD) or glycerol (YPG) and grown at 30°C. (B) The COX and cytochrome bc1 relative specific enzyme activities were determined and expressed as a percentage of the value for the wild-type control. The averages of three replicate measurements are given. (C) Serial 10-fold dilutions of wild-type (WT) cells and Δrcf1 Δrcf2 cells expressing Rcf1His or Rcf2His or not (−) were spotted on YP plates containing glucose (YPD) or glycerol supplemented with 0.1% galactose (YPG + 0.1% Gal) and grown at 30°C.
Affinity purification of His-tagged proteins.
Mitochondria (200 μg protein) were solubilized in lysis buffer (100 mM KCl, 20 mM HEPES-KOH, 10 mM MgCl2, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], pH 7.4), containing either 2.0% digitonin or 0.25% Triton X-100, or as otherwise indicated, for 30 min on ice and then Ni-NTA purification of His-tagged proteins was essentially performed as previously described (24).
Sucrose gradient centrifugation.
Mitochondria (200 μg protein) were solubilized in 20 mM potassium phosphate (pH 7.4), 15% glycerol, 2 mM EDTA, 0.25% Triton X-100, 1 mM PMSF, subjected to a clarifying spin, applied to a 5 to 50% sucrose (wt/vol) gradient, and centrifuged, essentially as described in reference 6. Fractions were collected, trichloroacetic acid (TCA) precipitated, and analyzed by SDS-PAGE and Western blotting.
Miscellaneous.
Chemical cross-linking with m-maleimidobenzoyl-N-hydroxysuccinimide ester (MBS) was performed essentially as described previously (25). NADH-cytochrome bc1 and COX enzyme activities were measured in the presence of sodium deoxycholate (0.5%) essentially as previously described (11, 24). The relative specific activities of the enzymes were calculated for both wild-type and mutant mitochondria and were averaged from three replica measurements. Mitochondrial isolation, protein determinations, carbonate extraction, blue native PAGE (BN-PAGE) and SDS-PAGE were performed according to published methods (2, 11, 24, 27). Translation in organello with [35S]methionine labeling was performed as described previously (24).
RESULTS
Rcf1 is associated with the cytochrome bc1-COX supercomplex.
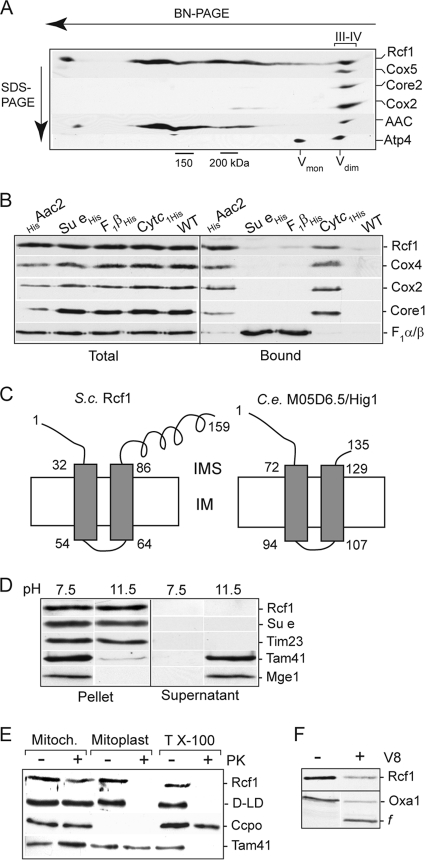
When analyzing the association of the Aac2 protein with the cytochrome bc1-COX supercomplex on native gels (BN-PAGE), we discovered a novel comigrating 18-kDa protein (Fig. 1A). Mass spectrometry analysis indicated it to be the protein encoded by the AIM31/YML030w gene, and it is referred to in this study as Rcf1. To directly demonstrate that the Rcf1 protein was physically associated with the cytochrome bc1-COX-AAC supercomplex (and not with the closely migrating F1Fo-ATP synthase dimeric complex), we performed an affinity purification of the cytochrome bc1-COX-AAC supercomplex using histidine (His)-tagged cytochrome c1 and Aac2 derivatives. Mild (digitonin) solubilization conditions were used to maintain the organization of this supercomplex (Fig. 1B). Rcf1 copurified with both cytochrome c1His and HisAac2 but not with His-tagged F1β and subunit e (Su e) proteins, two components of the dimeric ATP synthase complex. We conclude therefore that Rcf1, like the AAC proteins, exists in association with the cytochrome bc1-COX supercomplex.
Fig 1.
Rcf1, an inner membrane protein, associates with the cytochrome bc1-COX supercomplex. (A) Wild-type mitochondria were solubilized with digitonin (1%) and subjected to BN-PAGE electrophoresis followed by a second-dimension SDS-PAGE. The resulting Western blot was immunodecorated with antibodies directed against subunits of the COX (Cox5 and Cox2), cytochrome bc1 (Core2), and F1Fo-ATP synthase (Atp4) complexes, AAC, and Rcf1. The positions of the dimeric and monomeric ATP synthase (Vdim and Vmon), cytochrome bc1-COX supercomplex (III-IV), and 150- and 200-kDa markers are indicated. (B) Mitochondria were isolated from strains harboring either His-tagged Aac2, subunit e (Su e), F1β, or cytochrome c1 (Cytc1) proteins or from control wild-type cells (i.e., lacking a His-tagged protein), solubilized with digitonin (0.5%), and subjected to Ni-NTA purification, Western blotting, and immunodecoration. “Total” indicates 5% of the solubilized mitochondria; “Bound” indicates the Ni-NTA-eluted material. (C) Depiction of the Nout-Cout orientation of S. cerevisiae (S.c.) Rcf1 and C. elegans (C.e.) M05D6.5 (Hig1 homolog) in the inner membrane (IM), with N and C tails exposed to the intermembrane space (IMS). (D) Isolated wild-type mitochondria (0.1 mg/ml) were extracted by alkaline treatment using 0.1 M carbonate buffer at pH 11.5. Control mitochondria (pH 7.5) were resuspended in 0.6 M sorbitol, 20 mM HEPES-KOH, pH 7.5. Following extraction, samples were subjected to ultracentrifugation, and the resulting membrane pellet and supernatant (after TCA precipitation) were analyzed by SDS-PAGE, Western blotting, and immunodecoration for Rcf1 and control proteins. (E) Proteinase K (20 μg/ml) treatment was performed on intact mitochondria (Mitoch.) or mitochondria which had been subjected to hypotonic swelling (Mitoplast) or Triton X-100 detergent lysis (T X-100) on ice. Samples were subjected to SDS-PAGE and Western blotting. Cytochrome c peroxidase (Ccpo), a soluble IMS marker protein, displays inherent protease stability and hence is not degraded by the proteinase K treatment in the presence of Triton X-100. D-LD, d-lactate dehydrogenase, an IM protein with a large C-terminal domain exposed to the IMS. (F) Wild-type mitochondria were hypotonically swollen, subjected to treatment with V8 protease (20 μg/ml), and further processed as described for panel B. OxaI and the fragment (f) of OxaI resulting from V8 degradation of the N-tail of OxaI exposed to the IMS are indicated.
The Hig1 homology domain is localized to the N-terminal region of Rcf1, a region that encompasses two putative transmembrane segments. The C-terminal hydrophilic segment with a predicted coiled-coil domain is not a conserved feature of Hig1 proteins and appears to be limited to the fungal Rcf1 proteins (Fig. 1C). Carbonate extraction experiments demonstrated that Rcf1 was resistant to the alkaline extraction and thus behaved like the integral inner membrane control proteins, Su e of the Fo-ATP synthase and Tim23. In contrast, Mge1 and Tam41, which are matrix-soluble and matrix-localized peripheral inner membrane control proteins, respectively, were extracted by the carbonate treatment (Fig. 1D). Protease (proteinase K and V8 proteases) accessibility studies demonstrated that Rcf1 is localized in the inner membrane, where it became accessible to proteases only after hypotonic swelling (Fig. 1E and F). In mitoplasts, Rcf1 displayed a degree of susceptibility to the V8 protease similar to that of OxaI, a control inner membrane protein with an Nout-Cin orientation. As the V8 protease cleaves preferentially after negatively charged residues, found only in the N- and C-tail segments of Rcf1 (and not present in the hydrophilic loop between the two transmembrane domains of this protein), we conclude that the N and C termini of Rcf1 are exposed to the intermembrane space, where they are accessible to the V8 protease. Protease accessibility analysis of the C-terminal His epitope of Rcf1His also confirmed this membrane orientation (results not shown). In summary, we conclude that Rcf1 is an integral inner membrane protein that adopts an Nout-Cout orientation (Fig. 1C).
Rcf1 associates with Cox3 and partitions with the COX complex but also can bind to the cytochrome bc1 complex.
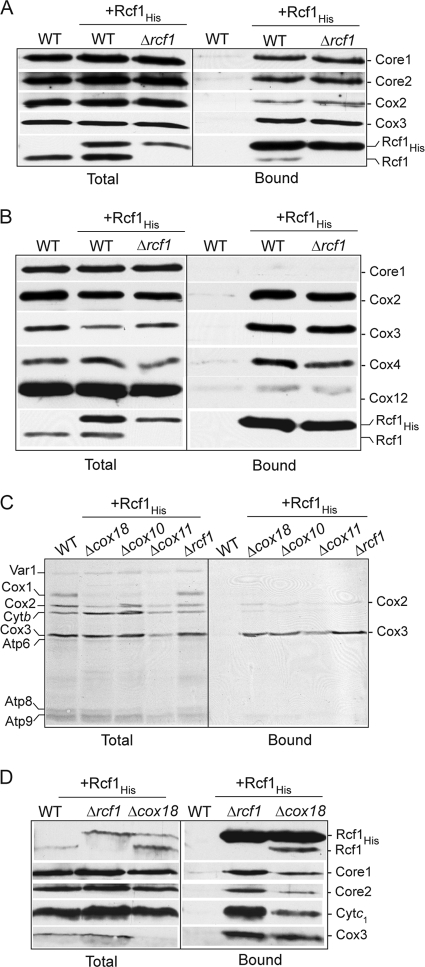
In order to further explore the association of Rcf1 with the cytochrome bc1-COX supercomplex, we expressed a C-terminally His-tagged Rcf1 in the wild-type and Δrcf1 strains. Under digitonin solubilization conditions, Rcf1His from both mitochondrial types was found in association with the cytochrome bc1-COX supercomplex, as demonstrated by the copurification of components of both the cytochrome bc1 (Core1 and Core2) and the COX (Cox2 and Cox3) complexes (Fig. 2A). To analyze if Rcf1 preferentially associates with either the cytochrome bc1 or COX aspect of this supercomplex, we next purified Rcf1His under Triton X-100 solubilization conditions, because this detergent causes the dissociation of the cytochrome bc1 from the COX complex (8, 24). Under these conditions, Rcf1His was observed to fractionate with the COX and not with the cytochrome bc1 complex, as indicated by the copurification of COX subunits and the absence of the Core1 protein (Fig. 2B). The efficiency of Cox12 copurification with Rcf1His was significantly less than that of the other COX subunits analyzed. Cox12 (equivalent to mammalian Cox6b [mCox6b]) is a peripheral COX subunit associated with the intermembrane-space side of the complex (4, 9, 23, 28, 41), which reflects the fact that the association of Cox12 with the COX complex is not as stable as other COX subunits under these Triton X-100 solubilization conditions (results not shown).
Fig 2.
Rcf1 can independently associate with Cox3 and the COX and cytochrome bc1 complexes. (A and B) Mitochondria isolated from wild-type (WT) cells and Δrcf1 cells harboring Rcf1His and control wild-type mitochondria were solubilized in 2% digitonin (A) or 0.25% Triton X-100 (B), subjected to Ni-NTA purification, and further analyzed as described for Fig. 1B. (C) In organello protein synthesis in the presence of [35S]methionine was performed in mitochondria isolated from wild-type control cells or from Rcf1His-containing cells. Following Triton X-100 solubilization, Ni-NTA purification was performed, followed by SDS-PAGE and autoradiography. (D) Rcf1His was purified from Δrcf1 or Δcox18 mitochondria under digitonin solubilization conditions and further analyzed as described for Fig. 1B.
To address the question of whether Rcf1 displayed the capacity to directly interact with one of the mitochondrially encoded cytochrome bc1 or COX subunits (cytochrome b, Cox1, Cox2, or Cox3), protein synthesis in the presence of [35S]methionine was performed in mitochondria isolated from the Δrcf1 strain harboring Rcf1His. Following translation, mitochondria were solubilized with Triton X-100, and the Rcf1His and associated proteins were affinity purified. Radiolabeled Cox3, and to a significantly lesser extent Cox2, was found to copurify with Rcf1His (Fig. 2C). As the assembly of newly synthesized Cox3 does not proceed to completion in isolated mitochondria, we conclude that the association of the Rcf1 and Cox3 proteins occurs independently of their final assembly into the COX enzyme. This conclusion was further supported by the observation that radiolabeled Cox3 synthesized in mitochondria isolated from the Δcox10, Δcox11, and Δcox18 mutants could also be affinity purified with Rcf1His (Fig. 2C). These mutant strains are defective in different stages of the COX assembly process, due to the absence of specific COX assembly factors (13, 30). In the absence of assembly into the COX complex, the Cox3 protein is susceptible to proteolytic turnover, but the low steady-state levels of Cox3 remaining in the isolated Δcox18 mitochondria were efficiently recovered in association with Rcf1His, under both Triton X-100 (results not shown) and digitonin (Fig. 2D) solubilization conditions.
We next took advantage of the COX assembly mutants to address the question of whether Rcf1 could associate with the cytochrome bc1 complex in the absence of an assembled COX complex. When Rcf1His was purified from Δcox18 mitochondria under digitonin solubilization conditions, Rcf1His was found in association, not only with Cox3, but also with the cytochrome bc1 complex, as evidenced by the coassociation of Core1, Core2, and cytochrome c1 proteins (Fig. 2D).
From these results, we conclude that Rcf1 is found together with the cytochrome bc1-COX supercomplex, where it must be supported by at least two independent associations, one contact point being with the cytochrome bc1 complex (which is digitonin stable but Triton X-100 sensitive) and the second being with the COX complex, which is Triton X-100 stable. Rcf1 associates with Cox3 prior to its assembly into the COX complex, and presumably this strong Rcf1-Cox3 interaction may contribute to the partitioning of Rcf1 with the assembled COX complex. Importantly, Rcf1 can interact with the Cox3 protein and cytochrome bc1 complexes independently of the assembly state of the COX complex.
The Rcf1 and Rcf2 proteins overlap in function and support the activity of the COX enzyme.
Deletion of the RCF1 gene in a haploid yeast strain did not significantly impact the respiration-based growth of the yeast cells, and weak growth of the Δrcf1 strain was observed on glycerol, a nonfermentable carbon source (Fig. 3A). Thus, Rcf1 does not play an essential role in the assembly of the COX (and/or cytochrome bc1 complex), as the activity of these enzymes is required for growth on glycerol media. BLASTp analysis indicated that Rcf1 is a member of a conserved family of proteins termed the hypoxia-induced gene 1 (Hig1) family (discussed further below), which bears limited similarity to a second yeast mitochondrial membrane protein, Aim38 (YNR018w) (22), referred to in this study as Rcf2. We therefore explored the possibility of genetic interaction between RCF1 and RCF2. Deletion of RCF2 alone failed to significantly impact the respiration-based growth of the haploid yeast. However, a yeast mutant deficient in both the Rcf1 and Rcf2 proteins, the Δrcf1 Δrcf2 strain, displayed a strongly impaired growth phenotype on nonfermentable media (Fig. 3A). The COX enzymatic activity levels were severely compromised in the resulting Δrcf1 Δrcf2 mitochondria and were present at approximately 20% of the levels in the wild-type control mitochondria. Deletion of either RCF1 or RCF2 alone only partially affected the COX enzymatic activity levels, with a stronger dependence on the Rcf1 protein being observed (Fig. 3B, top). The expression of Rcf1His fully restored the COX enzyme activity levels in the Δrcf1 Δrcf2 mitochondria, whereas the expression of Rcf2His resulted in only a partial recovery of the enzyme activity (Fig. 3B). As the COX levels in the Δrcf1 Δrcf2+Rcf2His mitochondria were not restored to those in the Δrcf1 (i.e., containing endogenous Rcf2) mitochondria, we conclude that the presence of the C-terminal His tag on the Rcf2 protein may have partially compromised Rcf2's stability and/or function. The cytochrome bc1 enzymatic activity levels were unaffected in the absence of Rcf1 (Fig. 3B, bottom). In contrast, elevated cytochrome bc1 activity levels were observed in the Δrcf2 mitochondria, suggesting that Rcf2 may function to exert control over the cytochrome bc1 enzyme. When both Rcf1 and Rcf2 were absent, the levels of cytochrome bc1 activity were partially affected suggesting that Rcf1 and Rcf2 proteins act differently to regulate this enzyme (Fig. 3B, bottom).
The expression of either the His-tagged Rcf1 or the Rcf2 derivative was observed to largely complement the respiratory growth defect of the Δrcf1 Δrcf2 mutant, where the His-tagged Rcf1 performed slightly better (Fig. 3C). Although the His-tagged Rcf2 protein failed to fully restore the COX enzyme levels in the Δrcf1 Δrcf2 mutant, the Rcf2His protein and COX activity were clearly present at levels sufficient to sustain respiration-based growth of the Δrcf1 Δrcf2 strain.
To confirm that Rcf2, like Rcf1, physically associates with the cytochrome bc1-COX supercomplex, we expressed Rcf2His in a Δrcf2 mutant and purified it under digitonin solubilization conditions. Subsequent silver stain analysis indicated that the cytochrome bc1-COX supercomplex copurified with Rcf2His (Fig. 4A), and was this confirmed with parallel Western blot analysis and immunodecoration for Core1 of the cytochrome bc1 complex and Cox2 and Cox3 of the COX complex (Fig. 4B). The level of cytochrome bc1-COX purified with Rcf2His was higher than with Rcf1His, and given that the levels of Rcf2His were lower than those of Rcf1His (as judged by the His epitope signal; as discussed above, this may indicate that the His epitope compromises that stability of Rcf2), it appears that the digitonin-solubilized Rcf2 protein can more efficiently (or more stably) associate with the cytochrome bc1-COX supercomplex than Rcf1 does. When purified under Triton X-100 conditions, Rcf2His was not recovered in association with either the cytochrome bc1 or the COX complex (results not shown), suggesting that its association with the supercomplex may require elements of both complexes and thus is stable only when the supercomplex remains together, i.e., under digitonin conditions, or simply that its association with the supercomplex is not Triton X-100 stable. In contrast to results obtained with Rcf1, we could not detect an association between newly synthesized Cox3 and Rcf2His even under milder digitonin solubilization conditions (results not shown). We cannot rule out the possibility that Rcf2 may also directly bind to the COX complex, possibly to the Cox3 subunit, but in a detergent-labile fashion.
Fig 4.
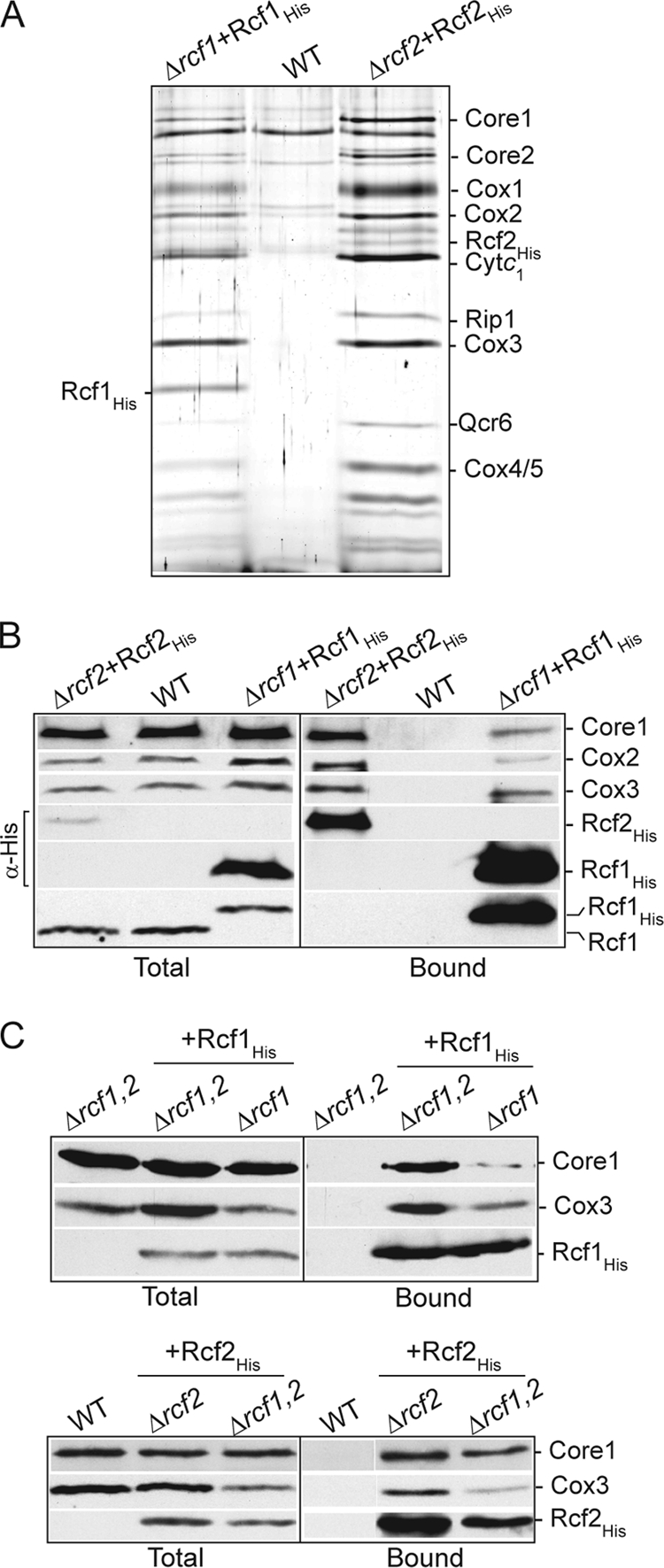
Rcf2, like Rcf1, associates with the cytochrome bc1-COX supercomplex. (A and B) Rcf1His and Rcf2His were expressed in their respective null mutant strains (Δrcf1+Rcf1His and Δrcf2+Rcf2His), and mitochondria were isolated and, together with wild-type control mitochondria analyzed in parallel, were solubilized in digitonin and subjected to Ni-NTA chromatography. Following SDS-PAGE of purified material, the gel was subjected to silver staining (A) or Western blotting and immunodecoration (B). (A) The identity of subunits verified by immunoblotting of parallel samples is indicated. (B) The levels of Rcf2His and Rcf1His were directly compared using an antibody against the His epitope (indicated by α-His). The presence of Rcf1 in the Rcf2His purified material was analyzed in a parallel sample using antibodies directed against the Rcf1 protein. Samples were further analyzed as for Fig. 1B. (C) Rcf1His (top) and Rcf2His (bottom) were purified under digitonin solubilization conditions from the indicated isolated mitochondria. Δrcf1,2 indicates Δrcf1 Δrcf2 mitochondria. Samples were further analyzed as described for Fig. 1B, and Cox3 and Core1 were immunodecorated. The recovery of Rcf1His and Rcf2His was monitored using an antibody directed against the His epitope.
Using His-tagged derivatives expressed in the Δrcf1 Δrcf2 strain, we could show that the association of either Rcf1 or Rcf2 with the cytochrome bc1-COX supercomplex did not depend on the presence of the other protein (Fig. 4C). These results confirm that Rcf1 and Rcf2 can independently associate with the cytochrome bc1-COX supercomplex. Consistently, the Rcf1 protein was not detected with the affinity-purified Rcf2His complex (Fig. 4A and B). (The lack of available antibodies against Rcf2, and the fact that Rcf2 would comigrate with the cytochrome c1 protein, makes it difficult at present to determine if the converse is true, i.e., that Rcf2 does not copurify with Rcf1His.) The independent association of Rcf1 and Rcf2 with the cytochrome bc1-COX supercomplex raises the interesting possibility that two different forms of this supercomplex, an Rcf1- and an Rcf2-associated one, may exist within mitochondria.
Taking all these results together, we conclude that Rcf1 and Rcf2 together play a critical role in the activity of the COX complex. The pronounced growth defect observed in the absence of both Rcf1 and Rcf2, and the ability of either Rcf1His or Rcf2His to complement both the growth phenotype and the COX activity defect observed in the Δrcf1 Δrcf2 mutant, demonstrates that Rcf1 and Rcf2 share overlapping functions. Consistent with their genetic interaction data, Rcf1 and Rcf2 can independently associate with the cytochrome bc1-COX supercomplex.
The assembly of the COX complex, and its association with the cytochrome bc1 complex, is altered in the absence of Rcf1 and Rcf2 proteins.
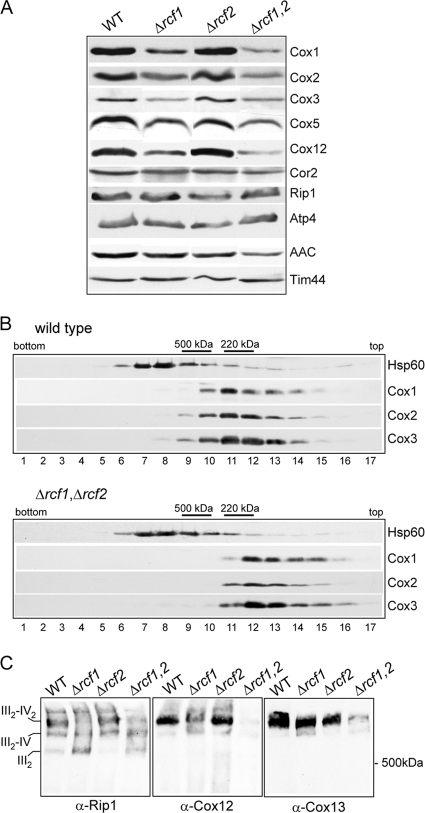
We next addressed the question of whether the reduced COX enzyme levels in the Δrcf1 Δrcf2 mitochondria was due to an altered assembly of the COX complex. The COX subunits (Cox1, Cox2, and Cox3 proteins) are highly susceptible to proteolytic turnover when their assembly is hindered, so the steady-state levels of these subunits can be used as an indicator of their assembly state. A partial reduction in the levels of these COX subunits was observed in the Δrcf1 mitochondria and to a similar extent in the Δrcf1 Δrcf2 mitochondria, indicating that the presence of Rcf1 was required for optimal stabilization of these proteins (Fig. 5A). However, given that the steady-state levels of these and other COX subunits (e.g., Cox5 and Cox12) were not significantly different between the single Δrcf1 mutant and the double Δrcf1 Δrcf2 mutant, we conclude that a simple reduction in the level of these COX subunits cannot be the underlying reason for the severely impaired COX enzyme activity observed when both Rcf1 and Rcf2 are absent. The levels of subunits from other OXPHOS enzymes, e.g., the Rieske Fe-S protein (Rip1) from cytochrome bc1 and Atp4 from the F1Fo-ATP synthase, were unaffected by the absence of Rcf1 and/or Rcf2. A reduction in the levels of the AAC protein was observed in the Δrcf1 Δrcf2 mitochondria, suggesting that the accumulation of these transport proteins may be influenced by the presence of the Rcf1 and Rcf2 proteins (Fig. 5A).
Fig 5.
The presence of Rcf1 and Rcf2 is required to secure the correct assembly of the COX complex and its association with the cytochrome bc1 complex. (A) Steady-state levels of the OXPHOS subunits in the mitochondria (50 μg) isolated from strains with the indicated genotypes. Δrcf1,2 indicates Δrcf1 Δrcf2 mitochondria. Tim44 was used as a loading control. Rip1, Rieske Fe-S protein. (B) Mitochondria isolated from WT and Δrcf1 Δrcf2 strains were solubilized in Triton X-100 and analyzed by sucrose gradient centrifugation, as described in Materials and Methods. The fractions corresponding to the bottom and top of the gradient are indicated. Following SDS-PAGE and Western blotting, immunodecoration was performed against the indicated COX subunits and Hsp60, a control marker protein. The positions of the control marker complexes monomeric ATP synthase (500 kDa) and cytochrome b2 (220 kDa) are indicated. (C) Mitochondria isolated from strains with the indicated genotypes were solubilized with digitonin (2%) and subjected to BN-PAGE analysis, Western blotting, and immunodecoration with Rip1, Cox12, and Cox13 antisera.
Knowing that Rcf1 can interact with newly synthesized Cox3, we next addressed the question of whether the presence of Rcf1 (and Rcf2) was required for the assembly of Cox3 with the core subunits of the COX complex. Mitochondrial proteins from the Δrcf1 Δrcf2 mutant and the wild-type control were solubilized with Triton X-100 and subjected to sucrose gradient centrifugation analysis (Fig. 5B). In the wild-type control, the COX subunits cofractionated with a size consistent with the monomeric enzyme (approximately 200 kDa). Cofractionation of Cox3 with the Cox1 and Cox2 proteins was also observed in the Δrcf1 Δrcf2 gradient, indicating that the assembly of Cox3 with the core COX subunits did occur in the absence of these Hig1 homologs. We did note that the COX complex in the Δrcf1 Δrcf2 gradient appeared to be slightly smaller, eluting one fraction closer to the top of the gradient than that in the wild type, pointing to a possible assembly defect of some peripheral COX subunits in the absence of the Rcf1 and Rcf2 proteins (Fig. 5B).
The importance of Rcf1 and Rcf2 for the assembly state of the cytochrome bc1-COX supercomplex was directly analyzed using digitonin solubilization and BN-PAGE analysis. Using antibodies against the Rieske Fe-S protein (Rip1) of the cytochrome bc1 complex, we observed that the assembly state of this supercomplex was adversely affected in the absence of Rcf1 (Fig. 5C, left). In wild-type mitochondria, as previously described, the majority of the cytochrome bc1 (III)-COX (IV) complex was present in its III2-IV2 form, whereas in the Δrcf1 sample, a significant amount of complexes corresponding to III2-IV and III2 forms were observed. The adverse effect on the assembly state of the cytochrome bc1-COX supercomplex was more pronounced in the Δrcf1 Δrcf2 mutant but not in the absence of Rcf2 alone. As the sucrose gradient analysis described above indicated that the assembly of peripheral COX subunits may be affected in the absence of Rcf1 and Rcf2, we used this BN-PAGE approach to directly analyze the assembly of the Cox12 and Cox13 proteins, two late-stage-assembled peripheral COX subunits (30). In the wild-type control mitochondria, the Cox12 and Cox13 proteins were found predominantly with the III2-IV2 form of the supercomplex. The assembly of Cox12 into the supercomplex was partially affected in the absence of Rcf1 alone and strongly defective in the Δrcf1 Δrcf2 double mutant (Fig. 5C, middle). The assembly of the Cox13 protein was also significantly reduced in the absence of Rcf1 and Rcf2 (Fig. 5C, right).
Taken together, our data demonstrate that the COX core subunits, including Cox3, can assemble in the absence of Rcf1 and Rcf2, but these hypoxia-induced gene 1 protein homologs are required to ensure the correct assembly of the cytochrome bc1-COX supercomplex. Furthermore, the association of the peripheral COX subunits Cox12 and Cox13, with the COX complex was perturbed, a defect that may underlie the observed compromised activity of the COX enzyme in the absence of both the Rcf1 and Rcf2 proteins.
The ability to support COX enzyme activity is a conserved feature of the Hig1 protein family.
The Hig1 homology domain of Rcf1 encompasses the N-terminal transmembrane region of the protein, whereas the extended C-terminal tail is found only in fungal Rcf1 proteins (Fig. 1C). In order to address the question of whether the conserved Hig1 homology domain of Rcf1 was responsible for the COX association, we created a His-tagged C-terminally truncated derivative (residues 1 to 112), Rcf1ΔCHis, where the C-terminal 47 residues were deleted. Although the steady-state levels of Rcf1 were compromised through this deletion, affinity purification experiments demonstrated that the Rcf1ΔC derivative retained the capacity to interact with the COX complex, as demonstrated by the copurification of the Cox1, Cox2, Cox3, and Cox4 subunits (Fig. 6A). From these data we conclude that the Hig1 homology domain of Rcf1 supports the COX association.
Fig 6.
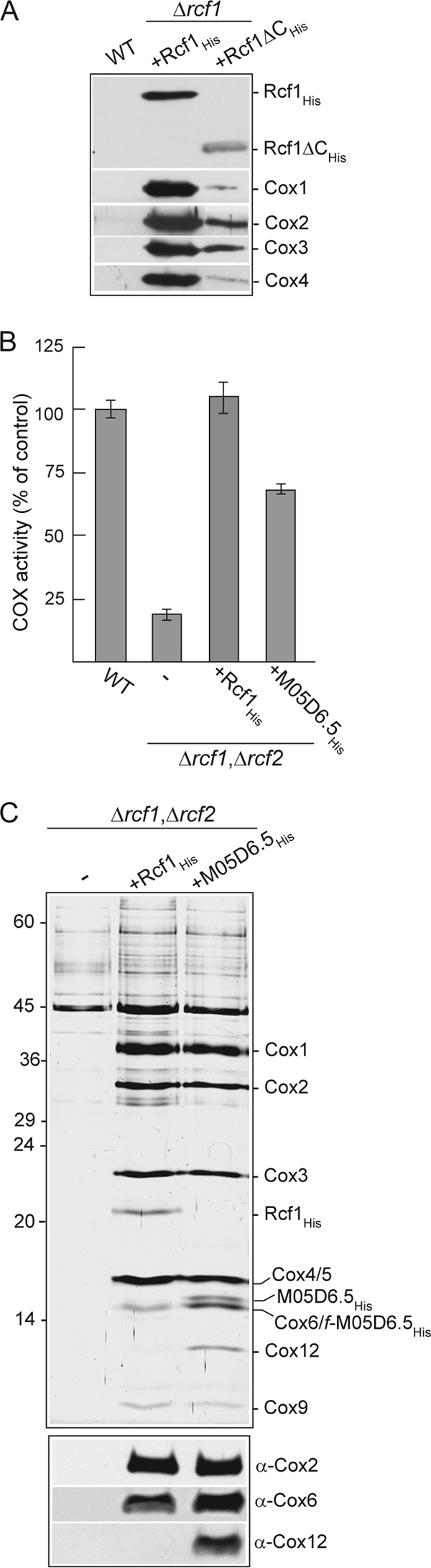
Physical association with and supporting activity of the COX enzyme are a conserved feature of Hig1 proteins. (A) Ni-NTA purification of Rcf1His and Rcf1ΔCHis following solubilization with Triton X-100. See Fig. 1B for further details. (B) COX enzyme activity measurements in mitochondria isolated from wild-type and Δrcf1 Δrcf2 strains harboring Rcf1His or the C. elegans Hig1 homolog M05D6.5His, as described for Fig. 3B. (C) Purification of Rcf1His and M05D6.5His from the Δrcf1 Δrcf2 strain performed under Triton X-100 solubilization conditions and analyzed by SDS-PAGE and silver staining (top) or in parallel by immunodecoration for Cox2, Cox6, and Cox12 subunits (bottom). The positions of COX subunits, Rcf1His, and M05D6.5His (and a smaller proteolytic fragment of this protein, f-M05D6.5His) on the silver-stained gel, verified by immunodecoration, are indicated.
To further demonstrate that the relationship with the COX complex was a conserved feature of the Hig1 proteins, we explored the question of whether a Hig1 homolog from the nematode C. elegans, the M05D6.5 gene product, could functionally substitute for the Rcf1 (and Rcf2) protein. The His-tagged derivative, M05D6.5His, was expressed in the Δrcf1 Δrcf2 strain, where it was observed to significantly restore the COX enzyme levels in the mitochondria (Fig. 6B). Furthermore, affinity purification of the M05D6.5His protein under Triton X-100 solubilization conditions demonstrated its ability to associate with the COX complex, as evidenced by the copurification of COX subunits (Fig. 6C, top). The level of the COX subunits associated with the M05D6.5His protein was similar to that with Rcf1His, with the notable exception of the Cox12 subunit. As was observed earlier (Fig. 2B), Cox12 did not efficiently copurify with Rcf1His (Fig. 6C, bottom); however, in contrast, Cox12 was clearly recovered with the purified C. elegans Hig1 homolog, M05D6.5His (Fig. 6C).
In summary, these data demonstrate that the ability to associate with and support the activity of the COX complex is not unique to the yeast Rcf1 and Rcf2 proteins but instead represents a conserved feature of the Hig1 protein family, and they carry out this association in a manner that appears to modulate the association of Cox12 with the COX complex.
The Rcf1 and AAC proteins exhibit a close physical relationship.
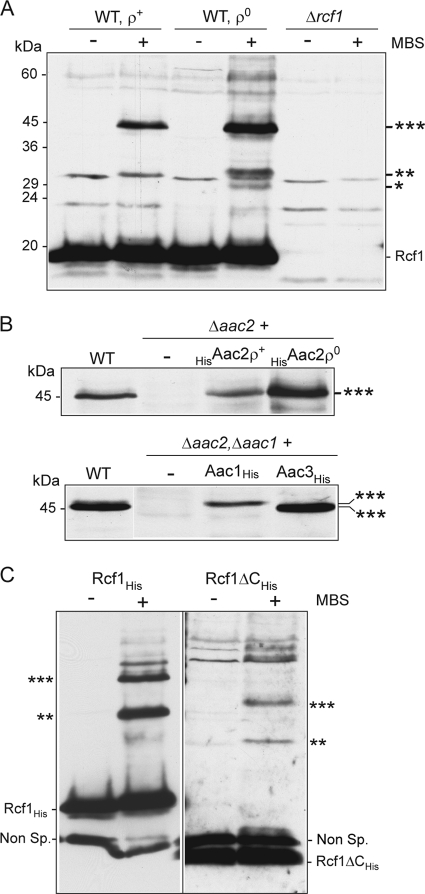
We used a chemical cross-linking approach to further probe the molecular environment of the Rcf1 protein. When the heterobifunctional (sulfhydryl/amino) reagent MBS was used, Rcf1 was found to form one dominant cross-link adduct of 45 kDa in wild-type mitochondria and corresponds to Rcf1 (18 kDa) and a partner protein of approximately 27 kDa. The ability of Rcf1 to form this adduct was not adversely affected and was even enhanced in [rho0] (i.e., mtDNA-deficient) mitochondria, demonstrating that this partnership was not dependent on the coassembly of Rcf1 with the cytochrome bc1-COX supercomplex, which is absent in [rho0] mitochondria. Furthermore, Rcf1 formed two less-abundant adducts of 33 kDa and 28 kDa, respectively, again more prominent in [rho0] mitochondria and corresponding to partner proteins of approximately 15 kDa and 10 kDa, whose identities are currently unknown (Fig. 7A).
Fig 7.
Rcf1 can be chemically cross-linked to AAC proteins. (A) Chemical cross-linking using MBS was performed on mitochondria isolated from wild-type (WT) [rho+] and [rho0] strains and the Δrcf1 strain. Following SDS-PAGE and Western blotting, immunodecoration with Rcf1 antiserum was performed. As the Rcf1 antibody recognizes some additional protein bands, the Δrcf1 mitochondria were analyzed in parallel to distinguish the Rcf1-specific adducts observed in WT mitochondria. ***, **, and *, 45-, 33-, and 28-kDa Rcf1-containing adducts, respectively. (B) Cross-linking of Rcf1 was performed in mitochondria isolated from the wild type and from the Δaac2 strain or the Δaac2 strain harboring HisAac2 protein in a [rho+] or [rho0] background (top) or from the Δaac2 Δaac1 strain harboring Aac1His or Aac3His (bottom). Samples were further analyzed as described for panel A. Only the area of the gel encompassing the 45-kDa Rcf1-containing adduct (***) is shown in both cases. (C) MBS cross-linking of Rcf1His and Rcf1ΔCHis was performed and further analyzed as described for panel A. The resulting Western blot was immunodecorated with antiserum which was directed against the His epitope but which also recognizes a nonspecific (Non Sp.) band of approximately 15 kDa. *** and **, Rcf1-containing adducts.
Given that the protein Aac2 is known to associate with the cytochrome bc1-COX complex (7, 11), we addressed the possibility that Aac2 and/or the other AAC isoforms could represent the partner of Rcf1 in the 45-kDa adduct. The ability of Rcf1 to form this adduct was lost in mitochondria isolated from the Δaac2 mutant strain. Expression of the HisAac2 derivative in the Δaac2 mutant restored the ability of Rcf1 to form this adduct, which again was enhanced when the experiment was performed in the Δaac2 HisAac2 [rho0] background (Fig. 7B, top). S. cerevisiae has three different AAC isoforms: the abundant Aac2 and the minor Aac1 isoform are constitutively expressed under aerobic conditions, while the Aac3 isoform is predominantly expressed under anaerobic conditions (33, 34). To test if Rcf1 could also cross-link to both the Aac1 and Aac3 isoforms, we performed cross-linking of Rcf1 in mitochondria from the Δaac2 Δaac1 mutant harboring His-tagged Aac1 and Aac3 proteins and expressed from a galactose-inducible promoter, GAL10. Rcf1 failed to form the 45-kDa adduct when cross-linking was performed in a Δaac2 Δaac1 mutant, but when the Aac1His or Aac3His isoform was expressed in this strain, the ability to form the adduct was restored and the size of the adduct was different: slightly larger when Aac1His was present and smaller when Aac3His was expressed (Fig. 7B, bottom). We conclude therefore that Rcf1 is physically close to the AAC isoforms within the mitochondrial membrane. The Rcf1ΔC protein also displayed the capacity to cross-link to the AAC protein, demonstrating that the association of Rcf1 with AAC proteins is a property of the Hig1 homology domain and not of the fungus-specific C-terminal domain of Rcf1 (Fig. 7C). No data to support a Rcf2-AAC interaction, however, have been obtained to date (results not shown), suggesting that the association with the AAC family members may be a specific property of the Rcf1 protein.
We conclude from these observations that the Hig1 homology domain of the Rcf1 protein interfaces with the AAC proteins and in a manner that is independent of Rcf1's or AAC's assembly with the cytochrome bc1-COX supercomplex.
DISCUSSION
We report here the identification of a novel component of the cytochrome bc1-COX supercomplex, the Rcf1 protein, which is a member of the conserved hypoxia-induced gene 1 (Hig1) family. Rcf1 was shown to genetically interact and share overlapping functions with the related Rcf2 protein, and both proteins can independently associate with the cytochrome bc1-COX supercomplex. We have chosen to term these Hig1 homologs, formerly referred to as Aim31 and Aim38 (22), the Rcf1 and Rcf2 proteins, or respiratory supercomplex factors 1 and 2, respectively. Our findings here indicate that the presence of Rcf1 and Rcf2 is particularly important for the enzymatic activity and correct assembly of the COX complex and its organization with the cytochrome bc1 supercomplex. Importantly, we demonstrate here that the C. elegans Hig1 homolog, M05D6.5, could substitute for the Rcf1/Rcf2 proteins in yeast, in its ability both to physically associate with the OXPHOS supercomplex and to support the enzyme activity of the COX complex. Thus, we conclude that their functional relationship with the respiratory supercomplex, and in particular with the COX enzyme, is an important and conserved feature of the Hig1 protein family members.
Although Rcf1 (formerly Aim31) and Rcf2 (formerly Aim38) were found in a genetic screen designed to identify mutants which display altered inheritance of mtDNA (22), our data indicate that the loss of COX enzyme activity in the absence of Rcf1 and Rcf2 proteins is not due to a possible loss of mtDNA and thus coding potential for the Cox1, Cox2, and Cox3 subunits. Cytochrome b, a key component of the cytochrome bc1 complex, is also encoded by the mtDNA, and the levels of this enzyme were not affected to the same extent as those of the COX complex in the absence of the Rcf1 and Rcf2 proteins and were even enhanced when Rcf2 alone was absent. Furthermore, in organello translation experiments showed that the capacity to synthesize Cox1, Cox2, Cox3 and cytochrome b was unaffected by the absence of Rcf1 and Rcf2 (results not shown). Thus, we conclude that the strong COX defect in the Δrcf1 Δrcf2 mutant is not indirectly due to limiting mtDNA levels but rather is directly due to the absence of Rcf1 and Rcf2 proteins and thus reflects the importance of these proteins for the correct assembly and function of the COX enzyme.
The Rcf1 and Rcf2 proteins represent novel cytochrome bc1-COX supercomplex components. Although both proteins are required to support the activity of the COX complex and although Rcf1, in particular, exhibits a strong affinity for the COX enzyme and Cox3 protein, we recognize that these proteins are unique as COX components because they display the ability to associate with the cytochrome bc1 complex in the absence of an assembled COX complex. Furthermore, the assembly of the cytochrome bc1-COX supercomplexes was altered when Rcf1 and Rcf2 were absent, consistent with the notion that these proteins may act as bridges by supporting both enzymes of the supercomplex. It is possible that the observed association of the Rcf1 protein with the cytochrome bc1 complex in the absence of COX complex assembly may be facilitated through Rcf1's association with Cox3, if Cox3 can associate with the cytochrome bc1 complex, prior to the completion of the COX assembly pathway. Further experimentation using Cox3 synthesis-defective mutants will be required to explore this possibility. On a separate note, it is important to point out that, while both Rcf1 and Rcf2 could be found in association with the cytochrome bc1-COX supercomplex, Rcf1 did not copurify with the Rcf2 protein, and our findings indicate that both proteins could independently associate with the OXPHOS supercomplex. These observations imply that at least two distinct populations of the cytochrome bc1-COX exist within the mitochondrial membrane system, an Rcf1-associated population and an Rcf2-associated one. The functional significance of these two different cytochrome bc1-COX supercomplex populations is currently unknown; however, one could speculate that their existence enables the regulation of the OXPHOS supercomplex through alternative mechanisms.
The demonstration here that the RCF1 and RCF2 genes genetically interact and that the Rcf1 and Rcf2 proteins can functionally substitute for each other is consistent with the documentation in the Saccharomyces genome database that Rcf2 (Aim38) displays a limited similarity to the Rcf1 (Aim31) and the Hig1 proteins and also agrees with an earlier report that these proteins cofractionated with the OXPHOS supercomplex on native gels (21). Our BN-PAGE analysis here, however, indicated that not all of the abundant Rcf1 protein was recovered with the cytochrome bc1-COX supercomplex and that a significant fraction of the protein was found in lower-molecular-mass complexes. Whether the smaller populations of the Rcf1 protein represent a proportion of the protein whose association with the cytochrome bc1-COX supercomplex was labile under the detergent conditions used, or whether a non-supercomplex-associated population of Rcf1 may exist in the mitochondria, is currently unclear. In contrast to traditional COX subunits, the levels of Rcf1 are not adversely affected in the absence of COX (or cytochrome bc1) assembly, e.g., in [rho0] mitochondria, an observation which may argue that a stable non-COX-associated Rcf1 population normally exists in the mitochondria. It is therefore possible that Rcf1/Hig1 proteins exhibit a dynamic association with the OXPHOS supercomplex that may serve to regulate the COX enzyme (and possibly cytochrome bc1) activities in response to certain metabolic cues.
We demonstrate here that Rcf1 physically associates with newly synthesized Cox3 prior to its assembly into the COX complex, suggesting that Rcf1 and the Hig1 proteins may represent Cox3-specific chaperones. Consistently, we observed that the steady-state levels of nonassembled Cox3 in the COX assembly mutants were elevated by increasing the levels of Rcf1, indicating that Rcf1 may serve to stabilize the nonassembled Cox3 protein (results not shown). On the other hand, our findings would suggest that Rcf1 does not represent an essential Cox3 chaperone in the traditional sense. First, the assembly of the Cox3 protein (as evidenced by its steady-state levels and coassociation of the COX core subunits Cox1 and Cox2) was not severely affected in the absence of Rcf1 (and Rcf2). Second, Rcf1 is retained with the COX complex following the assembly of Cox3, a behavior untypical of chaperones, which commonly dissociate from the relationship once the assembled state has been established. We propose that the observed association between Rcf1 and the unassembled Cox3 reflects a close physical and functional partnership between these two proteins, and one that is initiated before and continued after the assembly of the COX enzyme is complete. The demonstrated association with Cox3 enables us to map a potential site of contact between the Rcf1/Hig1 protein family members and the COX complex. The Cox3 subunit is conserved throughout prokaryotic and eukaryotic evolution, and although it does not directly contribute to the electron transfer and O2 reduction events, it is thought to play a supportive or regulatory role within the COX enzyme, possibly involving conformational changes in the Cox3 protein, and may be involved in adaptive changes to oxygen levels (32, 36, 41). The observed Rcf1-Cox3 relationship possibly creates a mechanism for Rcf1 to influence and regulate the activity of the COX enzyme, perhaps communicated through conformational changes in Rcf1 and/or Cox3 (or the associated cytochrome bc1 or AAC protein; see below). Cox3 together with Cox2 and Cox12 (mCox6b) defines the binding site for the enzyme's substrate, cytochrome c (9, 23, 41). In addition to Cox3, a number of our results here also point to a possible connection between Rcf1 and Cox12 subunit. First, the assembly of Cox12 into the cytochrome bc1-COX supercomplex was particularly affected in the Δrcf1 Δrcf2 mutant. Second, although the C. elegans Hig1 homolog M05D6.5His displayed a capacity similar to that of Rcf1His to associate with the COX enzyme, Cox12 was efficiently recovered with the purified nematode protein Hig1 but not with the yeast protein Rcf1. This observation may indicate that the Hig1 proteins function to influence the level or the nature of Cox12's association with the COX complex, possibly in a dynamic fashion, and that the C. elegans M05D6.5His protein displays an altered capacity in this respect, possibly due to the heterologous nature of the system. It is interesting in this respect that the phenotype of the Δrcf1 Δrcf2 mutant is reminiscent of the yeast cox12 null mutant; both contain a high level of the core COX subunits and display strongly reduced (about 15% of wild-type levels for Δcox12) yet measurable COX enzyme levels (28). Taking these results altogether, we suggest that the Hig1 proteins may serve to influence the substrate (cytochrome c) binding site of the COX complex, as defined by the arrangement of the Cox2, Cox3, and Cox12 (and possibly Cox13) proteins.
A close relationship between Rcf1 and the members of the AAC proteins family is also reported here. Thus, it is possible that Rcf1 is centrally positioned between the cytochrome bc1-COX supercomplex and the AAC proteins and that it functions to communicate between these complexes to regulate the activity of the OXPHOS enzyme in response to a perceived metabolic need of the cell. The ability of the Rcf1 to covalently cross-link to the AAC proteins was found to be differentially influenced by the specific inhibitors of the AAC enzymes atractyloside and bongkrekic acid (results not shown), inhibitors which each induce different conformations in the AAC proteins (33). Thus, we consider it possible that the Rcf1 protein may sense changes in the conformational states in the AAC protein and influence the activity of the COX complex through its ability to contact Cox3 and possibly the Cox2 and Cox12 proteins. Further investigation though is required to explore the proposed relationship between the AAC and Hig1 family members and their proposed influence on the cytochrome bc1 and/or COX enzymes.
The defining members of the Hig1 protein family were described in screens measuring genes that become upregulated under hypoxic conditions (1, 10, 19, 38, 42). Most Hig1 homologs documented in the databases since, however, have been identified through sequence comparisons to the founding family members, as the Hig1 proteins display a high degree of sequence conservation. Furthermore, in many cases multiple isoforms within the same species have been documented. Based on their sequence alignments, the Hig1 family members are subdivided into two distinct but closely related subgroups, the Hig1 type 1 and Hig1 type 2 subgroups. The Hig1 family members shown in the genetic screens to be upregulated under hypoxia and stress conditions predominantly belong to the Hig1 type 1 subgroup (1, 19, 10, 42), and from genome analysis this subgroup appears to be restricted to higher eukaryotes. We therefore consider it possible that the Hig1 type 2 subgroup may represent constitutively expressed isoforms, which function under nonstress conditions to support the COX complex activity and supercomplex stability. On the other hand, the Hig1 type 1 subgroup may represent isoforms expressed largely under hypoxic conditions, when it is necessary to assemble a COX complex optimized for operation under limiting O2 levels. In this respect, the regulation of the Hig1 type 2 and type 1 isoforms may be similar to that of the yeast Cox5a/5b (and mammalian Cox4-1/Cox4-2) system, where the Cox5b (mCox4-2) isoforms are expressed under hypoxic conditions and contribute to the assembly of a COX complex refined to operate under low O2 tensions (5, 7, 16, 26, 37). Interestingly, the AAC proteins, demonstrated here to exhibit a close physical relationship to Rcf1, also exist as multiple isoforms, one of which (Aac3 in yeast) is upregulated under hypoxic and anaerobic conditions (3, 18, 33, 34). It is important to note that no Hig1 type 1 homolog exists in S. cerevisiae or in C. elegans and that Rcf1, Rcf2, and M05D6.5 most closely resemble the Hig1 type 2 isoforms. Consistently, our preliminary data do not indicate a role for Rcf1 (or Rcf2) in supporting yeast growth under hypoxic or anaerobic conditions (results not shown). So the presence of a hypoxic-induced (type 1) isoform of Hig1 may be a feature limited to higher eukaryotes.
In summary, our data demonstrate that the Hig1 protein family members associate with the mitochondrial cytochrome bc1 and COX complexes, where they function to support the supercomplex organization and the activity of these enzymes, in particular the COX complex. Interestingly, putative homologs of the Hig1 proteins also exist in alphaproteobacteria, where their function is currently unknown. On the basis of our findings on the requirement of the Rcf1 and Rcf2 proteins for the assembly of an active COX enzyme, we propose that the Hig1 protein family, like the Shy1/SURF1 proteins (13, 30, 31), may represent evolutionarily important COX assembly and regulatory factors. Given that the Cox3 subunit, a primary contact point for Hig1 proteins in the mitochondrial COX, is conserved in bacteria, it is tempting to speculate that the prokaryotic Hig1 proteins also serve to interact with and modulate the activity of the COX enzyme through their Cox3 subunits.
ACKNOWLEDGMENTS
The cDNA clone encoding the C. elegans M05D6.5 Hig1 homolog was a kind gift from Yuji Kohara, Center for Genetic Resource Information, National Institute of Genetics, Mishima, Japan. We acknowledge and thank Mary Dienhart, Angela Tollefson, and Ru Ya for their contributions to this project. We are grateful to Klaus Pfanner, Freiburg, Germany, for the valuable gift of the Cox6, Cox12, and Cox13 subunit antisera.
The research was supported by NSF grants MCB 0744067 and NIH GM089699 to R.A.S.
Footnotes
Published ahead of print 6 February 2012
REFERENCES
- 1. Bedo G, Vargas M, Ferreiro MJ, Chalar C, Agrati D. 2005. Characterization of Hypoxia induced gene 1: expression during rat central nervous system maturation and evidence of antisense RNA expression. Int. J. Dev. Biol. 49: 431– 436 [DOI] [PubMed] [Google Scholar]
- 2. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248– 254 [DOI] [PubMed] [Google Scholar]
- 3. Brenner C, Subramaniam. K, Pertuiset C, Pervaiz S. 2011. Adenine nucleotide translocase family: four isoforms for apoptosis modulation in cancer. Oncogene 30: 883– 895 [DOI] [PubMed] [Google Scholar]
- 4. Carrero-Valenzuela RD, et al. 1991. Human cytochrome c oxidase subunit VIb: characterization and mapping of a multigene family. Gene 102: 229– 236 [DOI] [PubMed] [Google Scholar]
- 5. Castello PR, et al. 2008. Oxygen-regulated isoforms of cytochrome c oxidase have differential effects on its nitric oxide production and on hypoxic signaling. Proc. Natl. Acad. Sci. U. S. A. 105: 8203– 8208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Church C, Goehring B, Forsha D, Wazny P, Poyton RO. 2005. A role for Pet100p in the assembly of yeast cytochrome c oxidase: interaction with a subassembly that accumulates in a pet100 mutant. J. Biol. Chem. 280: 1854– 1863 [DOI] [PubMed] [Google Scholar]
- 7. Claypool SM, Oktay Y, Boontheung P, Loo JA, Koehler CM. 2008. Cardiolipin defines the interactome of the major ADP/ATP carrier protein of the mitochondrial inner membrane. J. Cell Biol. 82: 937– 948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cruciat C-M, Brunner S, Baumann F, Neupert W, Stuart RA. 2000. The cytochrome bc1 and cytochrome c oxidase complexes associate to form a single supracomplex in yeast mitochondria. J. Biol. Chem. 275: 18093– 18098 [DOI] [PubMed] [Google Scholar]
- 9. Das J, Miller ST, Stern DL. 2004. Comparison of diverse protein sequences of the nuclear encoded subunits of cytochrome c oxidase suggests conservation of structure evolving functional sites. Mol. Biol. Evol. 21: 1572– 1582 [DOI] [PubMed] [Google Scholar]
- 10. Denko N, et al. 2000. Epigenetic regulation of gene expression in cervical cancer cells by the tumor microenvironment. Clin. Cancer Res. 6: 480– 487 [PubMed] [Google Scholar]
- 11. Dienhart MK, Stuart RA. 2008. The yeast Aac2 protein exists in physical association with the cytochrome bc1-COX supercomplex and the TIM23 machinery. Mol. Biol. Cell 19: 3934– 3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dudkina NV, Kouril R, Peters K, Braun HP, Boekema EJ. 2010. Structure and function of mitochondrial supercomplexes. Biochim. Biophys. Acta 1797: 664– 670 [DOI] [PubMed] [Google Scholar]
- 13. Fontanesi F, Soto IC, Barrientos A. 2008. Cytochrome c oxidase biogenesis: new levels of regulation. IUBMB Life 60: 557– 568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fontanesi F, Clemente P, Barrientos A. 2011. Cox25 teams up with Mss51, Ssc1, and Cox14 to regulate mitochondrial cytochrome c oxidase subunit 1 expression and assembly in Saccharomyces cerevisiae. J. Biol. Chem. 286: 555– 566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fornuskova D, et al. 2010. Novel insights into the assembly and function of human nuclear-encoded cytochrome c oxidase subunits 4, 5a, 6a, 7a and 7b. Biochem. J. 428: 363– 374 [DOI] [PubMed] [Google Scholar]
- 16. Fukuda R, et al. 2007. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell 129: 111– 122 [DOI] [PubMed] [Google Scholar]
- 17. Gavriouchkina D, et al. 2010. Thyrotroph embryonic factor regulates light-induced transcription of repair genes in zebrafish embryonic cells. PLoS One 5: e12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Giraud S, Bono-Bidaud C, Wesolowski-Louvel M, Stepien G. 1998. Expression of human ANT2 gene in highly proliferative cells: GRBOX, a new transcriptional element, is involved in the regulation of glycolytic ATP import into mitochondria. J. Mol. Biol. 281: 409– 418 [DOI] [PubMed] [Google Scholar]
- 19. Gracey AY, Troll JV, Somero GN. 2001. Hypoxia-induced gene expression profiling in the euryoxic fish Gillichthys mirabilis. Proc. Natl. Acad. Sci. U. S. A. 98: 1993– 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Heinemeyer J, Braun H-P, Boekema EJ, Kouril R. 2007. A structural model of the cytochrome c reductase/oxidase supercomplex from yeast mitochondria. J. Biol. Chem. 282: 12240– 12248 [DOI] [PubMed] [Google Scholar]
- 21. Helbig AO, et al. 2009. A three-way proteomics strategy allows differential analysis of yeast mitochondrial membrane protein complexes under anaerobic and aerobic conditions. Proteomics 9: 4787– 4798 [DOI] [PubMed] [Google Scholar]
- 22. Hess DC, et al. 2009. Computationally driven, quantitative experiments discover genes required for mitochondrial biogenesis. PLoS Genet. 5: e1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hüttemann M, Jaradat S, Grossman LI. 2003. Cytochrome c oxidase of mammals contains a testes-specific isoform of subunit VIb—the counterpart to testes-specific cytochrome c? Mol. Reprod. Dev. 66: 8– 16 [DOI] [PubMed] [Google Scholar]
- 24. Jia L, Dienhart MK, Stuart RA. 2007. OxaI directly interacts with Atp9 and mediates its assembly into the mitochondria F1Fo-ATP synthase complex. Mol. Biol. Cell 18: 1897– 1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jia L, Kaur J, Stuart RA. 2009. Mapping the yeast OxaI-mitochondrial ribosome interface: identification of MrpL40, a ribosomal protein in close proximity to OxaI and critical for OXPHOS complex assembly. Eukaryot. Cell 11: 1792– 1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kwast KE, Burke PV, Staahl BT, Poyton RO. 1999. Oxygen sensing in yeast: evidence for the involvement of the respiratory chain in regulating the transcription of a subset of hypoxic genes. Proc. Natl. Acad. Sci. U. S. A. 96: 5446– 5451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680– 685 [DOI] [PubMed] [Google Scholar]
- 28. LaMarche AE, Abate MI, Chan SH, Trumpower BL. 1992. Isolation and characterization of COX12, the nuclear gene for a previously unrecognized subunit of Saccharomyces cerevisiae cytochrome c oxidase. J. Biol. Chem. 267: 22473– 22478 [PubMed] [Google Scholar]
- 29. Lenaz G, Genova ML. 2009. Structural and functional organization of the mitochondrial respiratory chain: a dynamic super-assembly. Int. J. Biochem. Cell Biol. 41: 1750– 1772 [DOI] [PubMed] [Google Scholar]
- 30. Mick DU, Fox TD, Rehling P. 2011. Inventory control: cytochrome c oxidase assembly regulates mitochondrial translation. Nat. Rev. Mol. Cell Biol. 12: 14– 20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mick DU, et al. 2007. ShyI couples Cox1 translational regulation to cytochrome c oxidase assembly. EMBO J. 26: 4347– 4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ogunjimi EO, Pokalsky CN, Shroyer LA, Prochaska LJ. 2000. Evidence for a conformational change in subunit III of bovine heart mitochondrial cytochrome c oxidase. J. Bioenerg. Biomembr. 32: 617– 626 [DOI] [PubMed] [Google Scholar]
- 33. Pebay-Peyroula E, Brandolin G. 2004. Nucleotide exchange in mitochondria: insight at a molecular level. Curr. Opin. Struct. Biol. 14: 420– 425 [DOI] [PubMed] [Google Scholar]
- 34. Sabová L, Zema I, Supek F, Kolarov J. 1993. Transcriptional control of AAC3 gene encoding mitochondrial ADP/ATP translocator in Saccharomyces cerevisiae by oxygen, heme and ROX1 factor. Eur. J. Biochem. 213: 547– 553 [DOI] [PubMed] [Google Scholar]
- 35. Schägger H. 2002. Respiratory chain supercomplexes of mitochondria and bacteria. Biochim. Biophys. Acta 1555: 154– 159 [DOI] [PubMed] [Google Scholar]
- 36. Scott GR, et al. 2011. Molecular evolution of cytochrome c oxidase underlies high-altitude adaptation in the bar-headed goose. Mol. Biol. Evol. 228: 351– 363 [DOI] [PubMed] [Google Scholar]
- 37. Semenza GL. 2007. Oxygen-dependent regulation of mitochondrial respiration by hypoxia-inducible factor 1. Biochem. J. 405: 1– 9 [DOI] [PubMed] [Google Scholar]
- 38. Shen C, Nettleton D, Jiang M, Kim SK, Powell-Coffman JA. 2005. Roles of the HIF-1 hypoxia-inducible factor during hypoxia response in Caenorhabditis elegans. J. Biol. Chem. 280: 20580– 20588 [DOI] [PubMed] [Google Scholar]
- 39. Stuart RA. 2008. Supercomplex organization of the oxidative phosphorylation enzymes in yeast mitochondria. J. Bioenerg. Biomembr. 40: 411– 417 [DOI] [PubMed] [Google Scholar]
- 40. Stuart RA. 2009. Supercomplex organization of the yeast respiratory chain complexes and the ADP/ATP carrier proteins. Methods Enzymol. 456: 191– 208 [DOI] [PubMed] [Google Scholar]
- 41. Tsukihara T, et al. 1996. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 Å. Science 272: 1136– 1144 [DOI] [PubMed] [Google Scholar]
- 42. Wang J, et al. 2006. Pancreatic β cells lack a low glucose and O2-inducible mitochondrial protein that augments cell survival. Proc. Natl. Acad. Sci. U. S. A. 103: 10636– 10641 [DOI] [PMC free article] [PubMed] [Google Scholar]