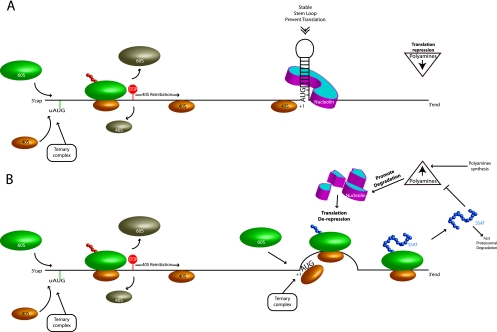
Fig 11.
Model of the SSAT translational control mechanism. (A) SSAT translational repression in the presence of low levels of polyamines. Success of ribosome scanning to the correct initiation codon is reduced in the presence of uORFs. Translation therefore depends on either leaky scanning or reinitiation of ribosome scanning 3′ to the uORFs. Those ribosome subunits escaping uORF control are then blocked by the strong stem-loop at the initiation site, which is stabilized by nucleolin. (B) Polyamine-regulated translational derepression of SSAT. An isoform of nucleolin blocks translation by binding to and stabilizing the stem-loop at the extreme 5′ end of the ORF. Increased polyamines cause autocatalysis of this nucleolin isoform, thereby reducing the stability of the stem-loop and allowing ribosomal subunits that escaped the uORF control to initiate translation. This increase in expression acetylates the polyamines and lowers their concentration, which reduces nucleolin degradation and thereby reestablishes translation repression. Together, these provide a negative feedback mechanism to regulate cellular polyamine concentrations.