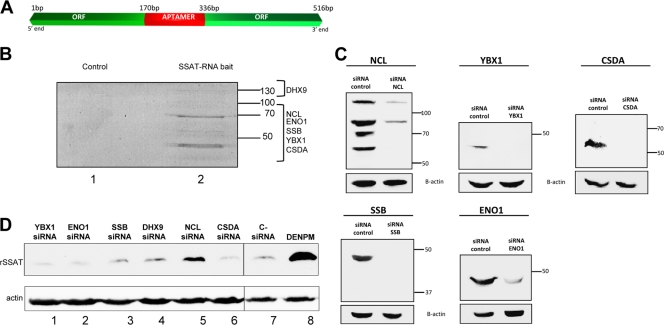
Fig 2.
SSAT RNA-interacting proteins and knockdown effects. (A) Chimeric RNA. Bait RNA was created using the first 170 bases of the 5′ end of the SSAT ORF and the last 181 bases of the 3′ end linked by a streptomycin-binding aptamer (red) that allowed the bait to bind to a Sepharose-streptomycin column. (B) Proteins interacting with SSAT RNA. HEK293T cell cytoplasmic proteins were incubated in the column, unbound proteins were washed away, and bound proteins were eluted with streptomycin-containing buffer. Eluted proteins were separated by SDS-PAGE and stained with Sypro ruby. No proteins were detected in the control column eluate, but multiple proteins were eluted from the SSAT-RNA column and then identified by mass spectroscopy (Table 1). Proteins and approximate gel positions are labeled: ENO1 (enolase 1), YBX1 (Y box protein 1), CSDA (DNA-binding protein A), SSB (La protein), DHX9 (ATP-dependent RNA helicase A), and NCL (nucleolin). (C) siRNA knockdowns. Western blots demonstrate individual siRNA knockdown of RNA-interacting proteins. (D) Knockdown effects on SSAT translation. Expression of recombinant SSAT was assayed by anti-His tag Western blotting with actin as the loading control. Lanes 1 to 6 are from cells treated with the indicated siRNA. Lane 7 is a negative control from cells treated with nontargeting siRNA. Lane 8 is a positive control with proteins from cells exposed to DENSPM to induce SSAT translation. Only nucleolin knockdown significantly increased expression of recombinant SSAT (lane 5).