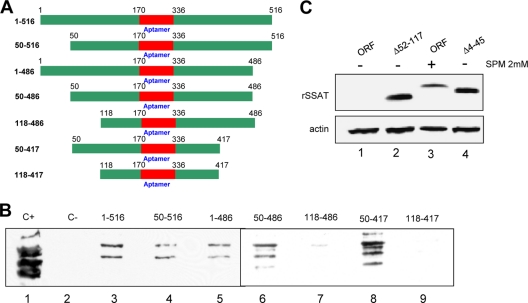
Fig 8.
SSAT RNA bases essential for nucleolin binding and translation repression. (A) Chimeric RNA-bait molecules. Top schematic, labeled 1-516, is the original chimera used to isolate nucleolin RNA binding proteins. Graphics and labels of others indicate nucleotides omitted in other bait molecules. (B) Nucleotides essential for nucleolin binding. Individual columns were prepared using the seven bait molecules plus a control with nonspecific RNA. HEK293T cell cytoplasmic lysate was incubated with the columns, and bound proteins were eluted and then concentrated for Western blot analysis using an antinucleolin monoclonal antibody. “C+” is a positive control using whole HEK293T lysate. “C-” is a negative control from the nonspecific RNA column. Lanes 3 to 9 are from columns prepared with the indicated bait molecules. All the columns bound nucleolin except those with baits lacking nucleotides 1 to 117. Since the 50–486 and 50–417 columns did bind nucleolin, binding depends on nucleotides within the 50-to-118 range. (C) Nucleotides essential for SSAT translational repression. HEK293T cells were transiently transfected with plasmids containing the complete SSAT ORF (ORF) and mutants using constructs lacking ORF nucleotides 52 to 117 (Δ52-117) or 4 to 45 (Δ4-45). Anti-His antibody was used for the blots, with antiactin for the loading control. Lanes 1 and 3 show that translation of the full SSAT ORF occurs only when the cells are exposed to spermine. Lanes 2 and 4 show that the Δ52-117 and Δ4-45 mutants are translated without spermine and thus both regions are required for translational control even if nucleolin binds to only one.