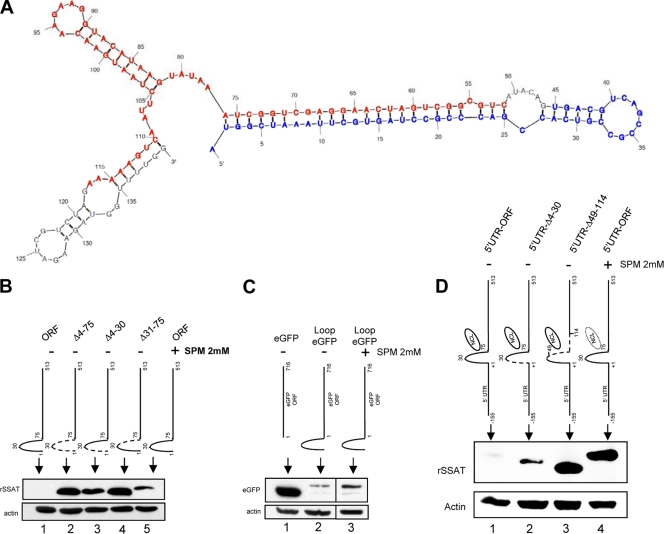
Fig 9.
SSAT RNA stem-loop and translational repression. (A) Mfold-calculated RNA secondary structure of first 140 bp of the SSAT ORF. Bases deleted in the Δ52-117 construct are marked in red; blue indicates the Δ4-45 construct. (B) Transiently transfected HEK293T cells. Transfecting plasmids contained the full SSAT ORF (ORF) or mutants with deletions that impair formation of a stem-loop by bases 2 to 76. The Δ4-75 mutant lacks the entire stem-loop, Δ4-30 lacks the first half, and Δ31-75 lacks the second half. Anti-His Western blots show that protein expression by the wild type is dependent on spermine exposure (lanes 1 and 5), but all mutants express protein without spermine (lanes 2, 3, and 4). (C) Fusion of the SSAT ORF stem-loop with a reporter. HEK293T cells were transiently transfected with plasmids producing RNA for the His-tagged reporter protein eGFP or RNA with the SSAT 1–75 stem-loop appended in frame to the 5′ end of eGFP (Loop eGFP). Anti-His Western blots show that Loop eGFP expression is repressed (lane 2) compared to that of eGFP (lane 1), and repression is partially relieved by exposure to 2 mM spermine (lane 3). (D) 5′ UTR and SSAT ORF stem-loop repression. To determine whether the 5′ORF stem-loop represses translation when the full 5′ UTR is present, HEK293T cells were transiently transfected with plasmids containing the 5′ UTR of SSAT RNA and the full coding region (5′UTR-ORF), the 5′ UTR with Δ4-30 ORF (5′UTR-Δ4-30), and the 5′UTR with Δ49-114 ORF (5′UTR-Δ49-114). Expression of 5′UTR-ORF required spermine exposure (lanes 1 and 4) while the mutants did not. Expression of 5′UTR-Δ49-114 (lane 3), which doesn't form the stem-loop and lacks the nucleolin binding region, was greater than that of 5′UTR-Δ4-30 (lane 2), which also doesn't form the stem-loop but does have the nucleolin binding region; the same relative relationship can be seen in panel B, although it is not as marked.