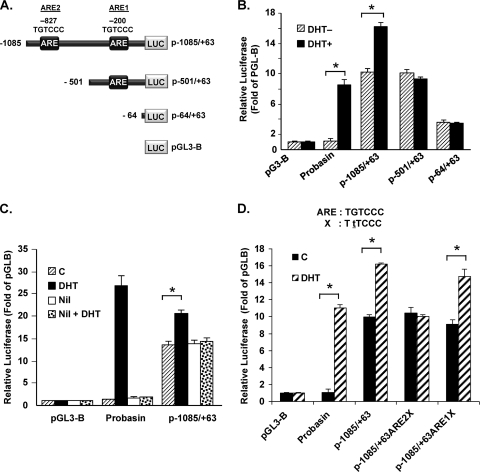
Fig 2.
Identification of functional ARE essential for DHT-induced GRTH promoter activity in GT1-7AR cells. (A) Schematic representation of GRTH-luciferase expression constructs p−1085/+63GL and 5′deletions. p−1085/+63GL comprises the GRTH minimal promoter region (bp −205/+63) relative to the ATG translational codon (+1) and 5′-flanking sequences with ARE half-sites located at bp −827 and −201 (boxes). (B) Serial deletion constructs indicated in panel A were transfected into GT1-7AR cells in the presence (black bars) or absence (striped bars) of DHT at 10−8 M. Luciferase activity is expressed relative to that of pGL3basic (empty vector). (C) DHT-induced promoter activity was also assessed in the presence or absence of nilutamide (Nil) treatment as described in the legend to Fig. 1. (D) Effect of a point mutation within the ARE half-site located at −827 or at −200 on DHT-induced promoter activity of p−1085/+63. WT ARE and the mutant form (X, with the mutated base underlined) are shown at the top. Luciferase activity is expressed relative to that of pGL3-B (empty vector) in all cases. The probasin construct (see Materials and Methods) was the positive control. Results are the means ± the standard errors of three individual experiments done in triplicate. *, P < 0.01.