Fig 3.
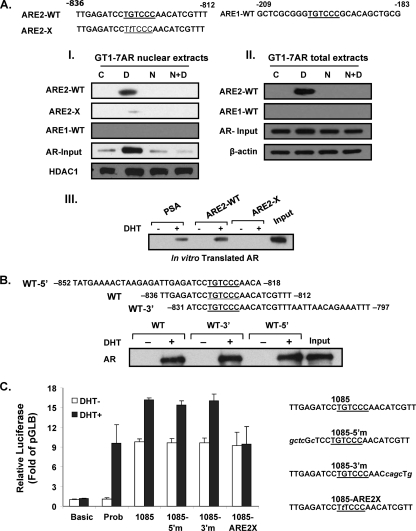
AR recruitment to the ARE half-site at −827/−822. DAPA were performed using probes for the WT ARE sites (underlined) with sequences 5′- and 3′-flanking nucleotides (10 bp). The ARE1-WT probe (−209/−183), the ARE2-WT probe (−836/−812), or a mutant form with a point mutation (G to T) in the ARE site at −826 (ARE2-X) was incubated with nuclear extracts (panel I) or total extracts (panel II) of GT1-7AR cells treated with the vehicle, DHT [D] (10−8 M), nilutamide [N] (1 μM), DHT plus nilutamide (N+D), or in in vitro-translated AR (panel III) in the presence or absence of DHT at 10−8 M. DNA-protein complexes were precipitated with streptavidin resin and subjected to Western blot analysis. Levels of AR are shown as the input using HDAC1 as a loading control for nuclear extracts or β-actin as a loading control for total extracts. Input, 10% in vitro-translated AR. Probasin (PSA), ARE2 WT (ARE2-WT), or mutant ARE2 (ARE2-X) was used as the probe. (B) Exploration of flanking sequence requirement for association of AR upon treatment with DHT to ARE2 located at bp −827/−822 of GRTH. DAPA analysis was performed using probes GRTH-WT (−836/−812), GRTH-WT-5′ (−852/−818), and GRTH-WT-3′ (−831/−797) with in vitro-translated AR in the presence or absence of DHT at 10−8 M. Ten percent in vitro-translated AR was included as the input. (C) Effects of mutations in 5′- or 3′-flanking sequence adjacent to ARE2 at −827 and sequences flanking nonfunctional ARE1 (italic) on DHT-induced promoter activity of p−1085/+63. WT p−1085/+63GL (1085) with the WT ARE (underlined) and mutations of the adjacent 5′ (5′m) and 3′ sequences (3′m) are shown on the right. Luciferase activity is expressed relative to that of pGL3 basic in all cases. The probasin construct (Prob) was used as a positive control. Results are the means ± the standard errors of three individual experiments done in triplicate.