Abstract
Early region 1A (E1A) of human adenovirus (HAdV) has been the focus of over 30 years of investigation and is required for the oncogenic capacity of HAdV in rodents. Alternative splicing of the E1A transcript generates mRNAs encoding multiple E1A proteins. The 55-residue (55R) E1A protein, which is encoded by the 9S mRNA, is particularly interesting due to the unique properties it displays relative to all other E1A isoforms. 55R E1A does not contain any of the conserved regions (CRs) present in the other E1A isoforms. The C-terminal region of the 55R E1A protein contains a unique sequence compared to all other E1A isoforms, which results from a frameshift generated by alternative splicing. The 55R E1A protein is thought to be produced preferentially at the late stages of infection. Here we report the first study to directly investigate the function of the species C HAdV 55R E1A protein during infection. Polyclonal rabbit antibodies (Abs) have been generated that are capable of immunoprecipitating HAdV-2 55R E1A. These Abs can also detect HAdV-2 55R E1A by immunoblotting and indirect immunofluorescence assay. These studies indicate that 55R E1A is expressed late and is localized to the cytoplasm and to the nucleus. 55R E1A was able to activate the expression of viral genes during infection and could also promote productive replication of species C HAdV. 55R E1A was also found to interact with the S8 component of the proteasome, and knockdown of S8 was detrimental to viral replication dependent on 55R E1A.
INTRODUCTION
Human adenovirus (HAdV) belongs to the family Adenoviridae. Viruses of this family have been isolated from vertebrates ranging from fish to humans. The first human adenoviruses were isolated in 1953 by independent groups searching for etiological agents responsible for acute respiratory infections (10, 21). However, it was not until 1962, when Trentin and colleagues discovered that HAdV-12 could cause tumors when injected into newborn hamsters, that interest in the field exploded (28). This was the first demonstration of a human virus that could cause cancer. Subsequent to that seminal observation, HAdV was used extensively to study fundamental biological processes ranging from regulation of the cell cycle to mRNA splicing.
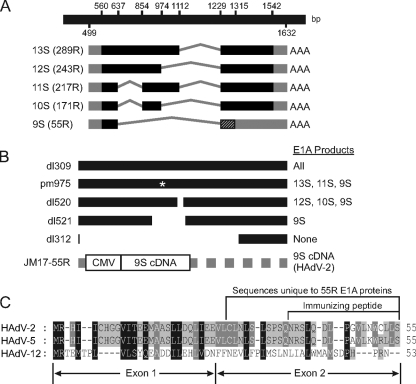
Later studies revealed that genes carried at the leftmost end of the HAdV genome were responsible for its oncogenic capacity. The early region 1A (E1A) transcript of HAdV type 2/5 (HAdV-2/5) encodes 5 proteins, of 289, 243, 217, 171, and 55 residues (R) (Fig. 1A). The mRNAs encoding these proteins are generated by differential splicing of a single primary RNA transcript. At the earliest stages of infection, E1A transcription is controlled by a constitutive enhancer, and expression of the largest two isoforms dominates, with the smaller three isoforms accumulating later (17, 27). While the largest two proteins have been studied extensively, no specific functions have been assigned to the smallest three isoforms. During infection, the major functions of the 289R and 243R E1A products include driving the host cell into S phase and activating the transcription of viral genes (2). These functions of E1A are essential for efficient replication of HAdV (12, 24).
Fig 1.
Structure of E1A transcripts, diagram of virus mutants, and 55R E1A protein alignment. (A) Graphical depiction of the mRNA species generated by splicing of the primary E1A transcript. (B) Graphical depiction of E1A region of viruses used in this study and the E1A proteins expressed by each virus. (C) Amino acid sequence comparison of the proteins encoded by the 9S mRNA species of HAdV-2, HAdV-5, and HAdV-12. Amino acids unique to 55R E1A and the peptide used for polyclonal antibody production are shown.
Recruitment of ATPase proteins independent of 20S (APIS) to early viral promoters by E1A enhances transcription of early viral genes (18). The HAdV-5 289R and 243R E1A proteins and the HAdV-12 266R and 235R E1A proteins can be immunoprecipitated with proteasomes (7). Amino acids 4 to 25 mediate binding of E1A to members of the 19S regulatory proteasome, human S8 and S4. Binding inhibits the ATPase activity of this subunit, which correlates with decreased proteasome activity (29). S8 is also recruited by conserved region 3 (CR3) to enhance transcription of early viral genes. Interestingly, the 20S proteasome is also recruited to CR3 independently of APIS and the 26S proteasome. E1A, S8, and the 20S proteasome are found on early gene promoters and sequences during infection and thus may be important in transcriptional initiation and elongation. In addition, inhibition of proteasome activity represses E1A-dependent transcriptional activation, further supporting the importance of this interaction during infection (18).
Despite the fact that no specific functions have been assigned to the smallest three E1A proteins, the 55R isoform constitutes a particularly interesting case. Its function has remained elusive despite the initial discovery of its mRNA species over 30 years ago (3). The 9S mRNA species that encodes the 55R E1A isoform accumulates preferentially at late times postinfection as the result of alterations in splice site usage (30). This accumulation seems to require replication of the viral genome. Indeed, the 9S mRNA is not produced in HEK293 cells unless they are infected with HAdV, despite the integration of genomic E1A and constitutive expression of 289R and 243R E1A in this cell line (4, 25, 26, 32, 37). The general lack of information about the 55R E1A isoform may be attributed, at least in part, to the fact that no existing E1A antibodies (Abs) are able to recognize the 55R product. While the 55R isoform shares the first 26 amino acids encoded by exon 1 with the other E1A proteins, reconstitution of the splice junction linking exons 1 and 2 causes a frameshift relative to the other isoforms, and this results in a unique C-terminal amino acid sequence (5, 19, 32) (Fig. 1A and C). These properties suggest that in addition to possibly binding targets that are known to interact with the extreme N terminus of the larger E1A isoforms, 55R E1A may also interact with unique targets via its novel C-terminal region. Due to the late kinetics of 55R E1A expression, these interactions may play important roles during the latest stages of HAdV infection.
In this study, a rabbit polyclonal antibody which specifically recognizes the 55R E1A protein encoded by HAdV-2 was generated and characterized. This antibody can be used for detection of HAdV-2 55R E1A by Western blotting, indirect immunofluorescence assay, and immunoprecipitation. A series of phenotypic and functional properties associated with 55R E1A are reported for the first time. 55R E1A is expressed most abundantly at late times postinfection. It was found to be localized to both the cytoplasm and the nucleus and was sufficient to promote virus replication in contact-inhibited IMR-90 fibroblasts. This may be due, in part, to the ability of 55R E1A to activate transcription of viral genes with kinetics and magnitudes that are unique in comparison to those for genomic E1A. Finally, a direct interaction of 55R E1A was observed with the APIS component S8 but not with S4. This is the first reported cellular target of 55R E1A. Knockdown of S8 was detrimental to virus replication, suggesting that this interaction is functionally important during infection.
MATERIALS AND METHODS
Cells and viruses.
Human embryonic kidney 293 (HEK293), HEK293T, HT1080, U2OS, and IMR-90 cells were originally obtained from the American Type Culture Collection (ATCC). dl309 (expresses all E1A proteins) (12), dl312 (does not express any E1A proteins) (12), dl520 (does not express 289R and 217R E1A) (8), dl521 (expresses only 55R E1A) (8), pm975 (does not express 243R and 171R E1A) (15), and HAdV-2 have all been described previously. JM17-55R was constructed by cloning the 55R E1A coding sequence into pXC1 (a generous gift of F. Graham). Recombinant virus was rescued by transfecting 5 μg of a new plasmid, pXC-55R HAdV-2 E1A, into HEK293 cells along with 10 μg of pJM17, using a 1:14 DNA-to-Superfect (Qiagen) ratio. Virus was then plaque purified and screened by sequencing of viral DNA (Fig. 1B). All cells were propagated in Dulbecco's modified Eagle's medium (DMEM) (Wisent) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin (all from Gibco). All viruses were grown on either HEK293 or A549 cells and were purified using a cesium chloride gradient as described previously (39).
Cell transfections and infections.
For virus replication assays, IMR-90 cells were seeded in 6-well dishes and were contact inhibited for 3 days after reaching confluence. For single-virus infections, cells were infected with either dl309, pm975, dl520, dl521, or dl312 at a multiplicity of infection (MOI) of 5 and were then incubated at 37°C and 5% CO2 for 1 h to permit adsorption. Cells were washed 5 times with PBS and were reincubated with fresh medium. For growth assays, supernatants were collected at 4, 48, and 120 h postinfection (hpi), and the titer of cell-free virus was assessed by plaque assay on HEK293 cells. For analysis of viral gene expression, cells were collected at 24, 48, 72, 96, 120, and 144 hpi. Virus coinfection replication assays were performed in a similar manner, with growth-arrested IMR-90 cells being infected with either pm975 plus dl521, pm975 plus dl312, dl520 plus dl521, or dl520 plus dl312 at an MOI of 5 for each virus.
For transfections, HT1080 and HEK293T cells were seeded on glass coverslips at a density of 5 × 104 cells/cm2 1 day prior to transfection with Superfect (Qiagen) according to the manufacturer's guidelines.
Small interfering RNA (siRNA) knockdown of S8 was performed on A549 cells seeded at 5 × 104 cells/cm2, using siLentFect (Bio-Rad) transfection reagent and 10 nM PMSC5 Silencer Select siRNA (Ambion). A second set of cells were treated with siRNA control 2 (Ambion). Following 12 h of incubation, cells were infected with either dl521 or dl312. Supernatants were collected at 24 and 96 hpi, and virus yields were determined by plaque assay on HEK293 cells.
mRNA isolation and qRT-PCR.
Total RNA was isolated using TRIzol reagent (Invitrogen). Contaminating viral DNA was eliminated by DNase I treatment (Invitrogen) according to the manufacturer's instructions. RNA was subjected to first-strand cDNA synthesis using Superscript II reverse transcriptase (Invitrogen) and a mixture of random hexamers and oligo(dT)20 according to the manufacturer's instructions. Quantitative reverse transcription-PCR (qRT-PCR) was performed using Express SYBR GreenER qPCR supermix (Invitrogen) on a Bio-Rad iQ5 iCycler according to the manufacturers' guidelines. E1B 2.2 kb, E3A, E4orf6/7 (18), and hexon (13) were amplified using previously described primers. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was amplified using the forward primer CCTGGCCAAGGTCATCCATGAC and the reverse primer TGTCATACCAGGAAATGAGCTTG. Conventional RT-PCR was performed using PCR-EZ D-PCR master mix (Bio Basic Inc.).
Plasmid construction.
pLE-9S and pEGFP-N1 were kind gifts of E. Moran and J. Torchia, respectively. All ligations were performed using T4 ligase (NEB) according to the manufacturer's instructions. To construct pCANmycEGFP-55R, HAdV-2 55R E1A-EGFP was cut from a preexisting vector, and the insert was ligated in frame with the myc epitope tag present in pCANmyc, which had also been blunted using the Klenow fragment. pEGFP-N1-55R was constructed by cloning the 55R E1A coding sequence in frame with the enhanced green fluorescent protein (EGFP) tag. pCANmyc-55R was constructed by cloning HAdV-2 55R E1A in frame with the N-terminal myc tag of pCANmyc. Glutathione S-transferase (GST)–55R/53R E1A fusions were made by cloning the 55R E1A proteins from HAdV-2 and HAdV-5, as well as the 53R E1A protein of HAdV-12, into the EcoRI and SalI sites of pGEX-4T1. pCDNA4-HA-S8 and pCDNA4-HA-S4 have been described previously (18).
Generation of anti-55R E1A antibodies.
Polyclonal Abs were generated against a peptide corresponding to the unique C-terminal region of HAdV-2 55R E1A: KYG-43-NRSLQDLPGVLNWCLLS-55. The peptide was coupled to keyhole limpet hemocyanin (KLH) by using bis-diazobenzidine (5 mg of peptide was conjugated to 5 mg of KLH). The KLH-peptide conjugate was injected subcutaneously into female New Zealand White rabbits. Each rabbit was injected at four sites with 100 μg at each site. Before injection, the KLH-peptide conjugate was emulsified with Freund's complete adjuvant for the initial inoculation and with Freund's incomplete adjuvant for subsequent injections. The rabbits were injected at 3-week intervals, and test bleeds were taken 10 days following the preceding injection.
Antibody reacting against the peptide was affinity purified using a peptide column that was prepared by conjugating 5 mg of peptide to a 6-ml bed volume of Affi-gel 10 (Bio-Rad) via hydroxysuccinimide linkage. The serum was diluted 2-fold in Tris-buffered saline (TBS) and then passed twice over the affinity column. The column was washed and then eluted with 100 μM glycine, pH 2.2, and the eluted antibody was dialyzed against TBS. The antibody was initially tested against sequential dilutions of the peptide spotted onto nitrocellulose. The rabbits underwent a total of eight injections.
Protein purification.
pGEX4T1-HAdV-2-55R, pGEX4T1-HAdV-5-55R, and pGEX4T1-HAdV-12 53R were expressed in Escherichia coli RIL (Stratagene) and were purified using standard methods.
Immunoprecipitation, GST pulldown, and immunoblot analyses.
For immunoprecipitation experiments, HEK293T or A549 cells were lysed in NP-40 lysis buffer (0.5% NP-40, 50 mM Tris, pH 7.8, 150 mM NaCl) supplemented with protease inhibitor cocktail (Sigma). One microgram of anti-GFP monoclonal Ab (MAb) (Clontech) was used for immunoprecipitation of EGFP-55R E1A, in combination with 125 μl of 10% protein A Sepharose resin (Sigma), from 0.5 mg of cell lysate. Samples were agitated for 1 h at 4°C. Beads were washed five times with lysis buffer, and samples were boiled in 1× lithium dodecyl sulfate (LDS) sample buffer for 5 min. Samples were separated in a sodium dodecyl sulfate (SDS)-polyacrylamide gel and were transferred onto a polyvinylidene difluoride (PVDF) membrane (GE Healthcare). Membranes were blocked in 5% nonfat milk in 1× Tris-buffered saline with 0.1% Tween 20.
For Western blots, cells were lysed in NP-40 lysis buffer and then boiled in sample buffer and treated as described above. Membranes were stripped by heating in a 2 M glycine buffer, pH 2.2, with 0.5% SDS. Ponceau staining was performed according to standard protocols.
Dot blot assays were performed according to standard procedures. Briefly, lysates from A549 cells infected with HAdV-2 or JM17-55R at an MOI of 10 were prepared under nonreducing conditions. Five-microgram aliquots of lysates were spotted onto a PVDF membrane and were probed with either M73 or anti-55R E1A polyclonal antiserum.
GST pulldown assays were performed using 0.25 μg of GST-55R E1A and 0.5 mg of lysate from HEK293T or A549 cells that had been transfected with constructs expressing hemagglutinin-S8 (HA-S8) or HA-S4 or were left untransfected. Samples were agitated for 1 h at 4°C with 12.5 μl of a 50% glutathione Sepharose slurry and were then treated as described for immunoprecipitation experiments. HA-S8 and HA-S4 were detected using rat anti-HA MAb (1:2,000) (3F10; Roche). EGFP was detected using anti-GFP MAb (1:2,000) (Clontech). 55R E1A was detected using custom rabbit polyclonal anti-HAdV-2 55R E1A antibodies (1 μg/ml). Input GST-tagged proteins were detected by Coomassie blue staining.
Rabbit polyclonal anti-S8 has been described previously (33). Secondary antibodies used included goat anti-mouse IgG (1:200,000) (Jackson Laboratory), goat anti-rabbit IgG (1:200,000) (Jackson Laboratory), and goat anti-rat IgG (1:20,000) (Pierce); all were conjugated to horseradish peroxidase. Membranes were incubated with ECL+ substrate (GE Healthcare) for 1 min prior to exposures.
Immunofluorescence microscopy.
All cells were seeded on coverslips in 24-well tissue culture dishes and were fixed in 3.7% paraformaldehyde (Fisher) for 30 min at room temperature. After washing in phosphate-buffered saline (PBS), cells were permeabilized on ice using 0.2% Triton X-100 (Biobasic) for 20 min. Coverslips were transferred to humidity chambers and were blocked using 10% FBS and 1% goat serum in PBS (blocking buffer [BB]) for 30 min at room temperature. Cells were incubated at room temperature for 1 h with anti-55R E1A rabbit polyclonal Abs (1:50) and/or anti-myc (9E10 hybridoma supernatant, used undiluted) primary Ab. After washing three times with BB, the cells were incubated for another hour at room temperature with Alexa Fluor 546-conjugated goat anti-mouse IgG, Alexa Fluor 594-conjugated goat anti-rabbit IgG, and/or Alexa Fluor 488-conjugated goat anti-mouse IgG (all from Molecular Probes). Finally, cells were washed three times with PBS, and nuclei were labeled with 0.2 mg/ml Hoechst 33342 (Molecular Probes) for 3 min at room temperature or with propidium iodide (25 μg/ml) for 30 min at room temperature. Cells were washed three more times, and coverslips were then mounted on glass microscope slides, using mounting medium consisting of 90% glycerol (Biobasic), 10% PBS, and 2.5 g/liter 1,4-diazabicyclo (2,2,2)octane (DABCO; Alfa Aesar). Imaging was performed using a Zeiss Axioskop 2 magneto-optical trap fluorescence microscope equipped with a QImaging Retiga 1300-coded monochrome 12-bit camera. Images were captured and pseudocolored using Northern Eclipse software, version 7.0. Confocal images were acquired using a Zeiss LSM 510 Meta confocal laser scanning microscope equipped with Zeiss Zen imaging software for analysis.
RESULTS
Characterization of anti-55R E1A polyclonal Abs.
Despite identification of an mRNA encoding the putative 55R E1A product of species C HAdV over 30 years ago, the protein itself has never been detected or systematically characterized in the context of infection. This is likely related to the limited sequence overlap with other E1A isoforms, such that none of the existing Abs which recognize various epitopes of E1A from species C HAdV are able to detect the 55R isoform. To address this issue, rabbit polyclonal antibodies were generated that specifically recognize the 55R E1A species of subgroup C HAdV.
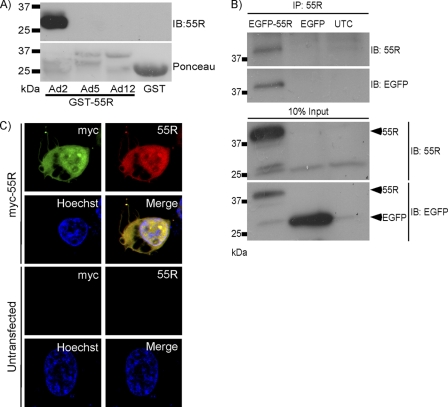
A peptide was synthesized to correspond to residues 43 to 55 of the unique C-terminal region of HAdV-2 (KYG-43-NRSLQDLPGVLNWCLLS-55) (Fig. 1C). This peptide was coupled to keyhole limpet hemocyanin and was used to immunize rabbits. The HAdV-2 55R E1A sequence was originally chosen because HAdV-2 was the first HAdV to be sequenced, and thus the 55R E1A cDNA clones that have been used previously were generated from HAdV-2. There are also very few differences between the 55R E1A sequences from HAdV-2 and HAdV-5. Abs were affinity purified from rabbit serum, and specificity was demonstrated by a dot blot assay using the immunizing peptide (data not shown). In order to determine the breadth of specificity of the affinity-purified Abs, a Western blot was performed to detect purified GST-55R E1A from HAdV-2 and HAdV-5, as well as the equivalent 53R E1A isoform of HAdV-12. The antibody detected 55R E1A from HAdV-2 but not from HAdV-5 (Fig. 2A), a closely related species C HAdV with only 3 nonidentical amino acids in the C terminus of the protein (Fig. 1C). The Abs were also unable to recognize the 53R E1A protein of HAdV-12, a more divergent species A HAdV, or GST alone (Fig. 2A).
Fig 2.
Characterization of 55R E1A antiserum. (A) Western blot and Ponceau staining of GST-purified 55R E1A (or 53R E1A, in the case of HAdV-12), obtained using rabbit polyclonal anti-55R E1A Abs. Upper bands of Ponceau staining correspond to E1A proteins. (B) EGFP-55R E1A was immunoprecipitated from HT1080 cells by use of rabbit polyclonal anti-55R E1A Abs. (C) Indirect immunofluorescence images of HT1080 cells expressing myc-55R E1A or left untransfected and stained with an anti-myc Ab and anti-55R E1A polyclonal Abs. Nuclei were stained with Hoechst 33342.
We next sought to determine whether the Abs could be used to immunoprecipitate HAdV-2 55R E1A. To do this, lysates were prepared from HEK293T cells expressing EGFP-55R E1A or EGFP alone or from mock-transfected cells. Lysates were then incubated with polyclonal anti-55R E1A Abs along with protein A Sepharose beads. After washing the beads thoroughly and boiling them with LDS sample buffer, proteins were separated by SDS-PAGE and were transferred to a PVDF membrane. A band corresponding to the molecular size of EGFP-55R E1A appeared only in the sample from EGFP-55R E1A-expressing cells when the membrane was probed with anti-55R E1A polyclonal antibodies or anti-GFP MAb (Fig. 2B). Thus, the polyclonal anti-55R E1A Abs likely recognize both native and denatured HAdV-2 55R E1A.
Finally, we determined whether our anti-55R E1A Abs could be used to study the subcellular localization of 55R E1A by indirect immunofluorescence assay. HT1080 cells were either mock transfected (data not shown) or transfected with a construct that expressed myc-55R E1A. At 24 h posttransfection, cells were fixed and stained using anti-55R E1A and anti-myc, followed by Alexa Fluor 594-conjugated goat anti-rabbit IgG or Alexa Fluor 488-conjugated goat anti-mouse IgG. Samples were analyzed by confocal microscopy, and colocalization of the signal was observed only in cells expressing myc-55R E1A (Fig. 2C), demonstrating that the Ab could also be used for determination of the subcellular localization of 55R E1A. 55R E1A could be found in both the nuclei and the cytoplasm of these cells. 289R and 243R E1A proteins have also been reported to be located in both the nucleus and cytoplasm, with the major pool of the proteins being nuclear (6, 9, 14).
55R E1A is expressed most highly late during infection and is localized to both the nucleus and the cytoplasm.
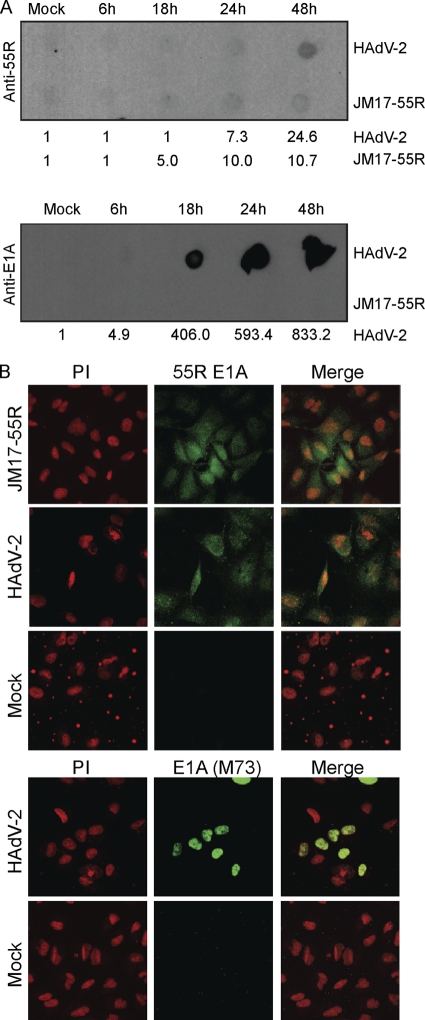
In order to elucidate the function of 55R E1A during HAdV infection, it was first important to determine the kinetics of 55R E1A protein expression and its subcellular localization. Expression kinetics of the 55R E1A protein were assessed by infection of A549 cells with HAdV-2 or JM17-55R at an MOI of 10. Cells were collected between 6 and 48 hpi (at which point cytopathic effects were evident in HAdV-2-infected cells). In cells infected with HAdV-2, the larger E1A isoforms could easily be detected by 18 hpi, with levels of protein accumulating for up to 48 hpi. In contrast, 55R E1A expression was evident at 24 hpi and increased approximately 3-fold by 48 hpi. 55R E1A expression became evident at 18 hpi in cells infected with JM17-55R (contains 55R E1A cDNA) and accumulated to 48 hpi, although expression was lower than that observed for HAdV-2 (Fig. 3A). This was likely due to stimulation of the E1A promoter by the 289R E1A product during HAdV-2 infection.
Fig 3.
Expression kinetics and subcellular localization of 55R E1A during HAdV-2 infection. Human A549 cells were infected with HAdV-2 or JM17-55R at an MOI of 10 or were mock infected. (A) Cells were collected at 6, 18, 24, or 48 hpi, and 5 μg of nonreduced lysate was blotted using anti-55R E1A polyclonal Abs or anti-E1A (M73). Numbers beneath blots are densitometry readings. (B) Subcellular localization of 55R E1A during infection was assessed at 48 hpi. Cells were stained with either anti-55R E1A polyclonal Abs or anti-E1A (M73), followed by Alexa Fluor 488. Nuclei were stained with propidium iodide (PI).
The subcellular localization of 55R E1A was assessed in A549 cells at 48 hpi. In both JM17-55R- and HAdV-2-infected cells, 55R E1A was localized diffusely in both the cytoplasm and the nucleus. As expected, the larger E1A proteins were found predominately in the nucleus. During HAdV-2 infection, chromatin organization was clearly altered in some cells. These morphological changes did not take place in cells infected with JM17-55R (Fig. 3B). Although the 55R E1A signal was weak at earlier time points, localization did not appear to change during the course of infection (data not shown).
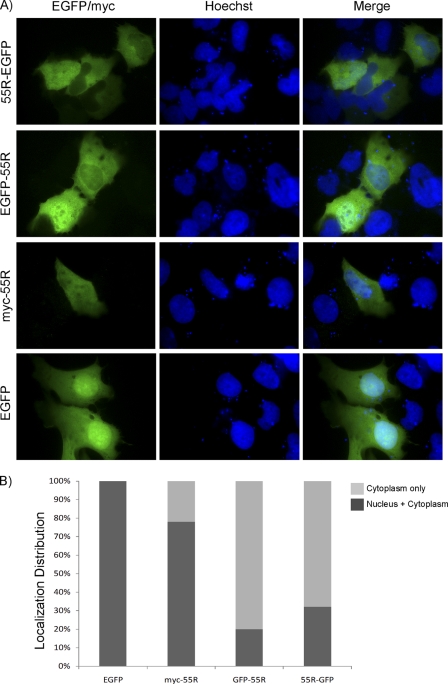
To more closely examine the subcellular localization of 55R E1A and the sequences of tagged 55R E1A constructs, as well as that of untagged 55R E1A, we performed an in silico analysis of subcellular localization. Untagged 55R E1A was predicted to be localized to the nucleus by SubLoc (11) and to the cytoplasm by virus-mPLoc (22, 23). These predictions were the same as those made for myc-55R E1A. Both EGFP-55R E1A and 55R E1A-GFP were predicted to have cytoplasmic localization by SubLoc, but virus-mPLoc predicted that EGFP-55R E1A would have a dual nuclear-cytoplasmic localization, whereas 55R E1A-EGFP was predicted to exhibit cytoplasmic localization only.
To empirically test these predictions, the constructs were transfected into U2OS cells (Fig. 4) and HT1080 cells (data not shown). As expected, EGFP was localized to both the nucleus and the cytoplasm in 100% of cells. myc-55R E1A exhibited dual nuclear-cytoplasmic localization in 78% of cells, whereas it was found primarily in the cytoplasm in 22% of cells. In cells expressing EGFP-55R E1A or 55R E1A-EGFP, the protein was found mainly in the cytoplasm in 80% and 68% of cells, respectively. Only 20% of cells expressing EGFP-55R E1A and 32% of cells expressing 55R E1A-EGFP exhibited dual nuclear-cytoplasmic localization of the respective fusion proteins (Fig. 4A and B). These experimental results matched well with the localizations predicted by the in silico analysis. No appreciable differences were observed when the constructs were expressed in HT1080 cells. Untagged 55R E1A could not be detected when it was expressed exogenously, possibly due to low expression levels and/or a shorter protein half-life than that observed during infection. Indeed, the GFP-tagged 55R E1A proteins could be detected in a much larger proportion of cells than myc-tagged 55R E1A, indicating that the large GFP tag may increase the half-life of the protein.
Fig 4.
Subcellular localization of 55R E1A. Human U2OS osteosarcoma cells were transfected with constructs encoding EGFP, myc-55R E1A, EGFP-55R E1A, or 55R E1A-GFP. (A) Nuclei were stained with Hoechst 33342, and myc-55R E1A was detected using monoclonal anti-myc (9E10) Ab followed by Alexa Fluor 488-conjugated goat anti-mouse IgG. (B) The subcellular localization of each construct was quantified by counting no fewer than 10 fields. Magnification, ×1,000.
55R E1A stimulates viral gene expression.
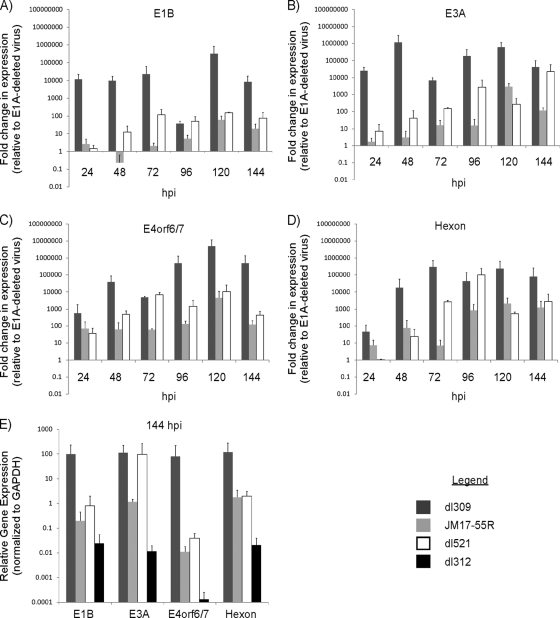
Given the localization of 55R E1A in the nucleus during infection, we sought to assess whether, akin to the largest E1A isoforms, 55R E1A could activate viral gene expression. To accomplish this, we performed a qRT-PCR assay on contact-inhibited human diploid IMR-90 fibroblast cells infected with dl309, JM17-55R, dl521, or dl312, using primers that recognize transcripts expressed from selected viral promoters (1, 13, 18). The E1A products expressed by each of these viruses are listed in Fig. 1B. RNA was extracted at 24, 48, 72, 96, 120, or 144 hpi. All samples were normalized internally to GAPDH, and viral gene expression was compared to that observed in samples from cells infected with dl312, a virus that lacks E1A. For these experiments, all transcript levels above a value of 1 can be attributed specifically to the presence of the E1A isoform(s) expressed by each virus.
dl309, which expresses all E1A isoforms, induced expression of E1B approximately 12,000-fold relative to that induced by dl312 (which does not express any E1A proteins) at 24 hpi. The level of activation induced by dl309 reached a maximum of approximately 317,000-fold by 120 hpi. For comparison, JM17-55R, which constitutively expresses only 55R E1A, resulted in only a 2.5-fold induction of E1B expression relative to that induced by dl312 at 24 hpi, which increased to a maximum of 61-fold induction at 120 hpi. In agreement with the known late expression kinetics of the mRNA encoding the 55R E1A protein, the levels of E1B steadily increased during dl521 infection (harbors genomic E1A and splices only 55R E1A), up to a maximum of 150-fold at 120 hpi (Fig. 5A). Thus, 55R E1A is able to induce expression of E1B to a modest extent compared to wild-type E1A. In addition, the maximal degree of induction remained consistent whether 55R E1A was expressed constitutively from a virus constructed using a 9S cDNA (JM17-55R) or from a virus in which splicing of the primary E1A transcript is required to generate the 9S mRNA (dl521).
Fig 5.
Influence of 55R E1A on viral gene expression. (A to D) Contact-inhibited primary IMR-90 fibroblasts were infected with dl309, JM17-55R, dl521, or dl312 at an MOI of 5. Cells were collected at 24, 48, 72, 96, or 120 hpi. Samples were subjected to qRT-PCR analysis using primers specific to transcripts controlled by various HAdV promoters, including E1B (A), E3A (B), E4orf6/7 (C), and hexon (D). (E) Relative expression levels at 144 hpi of viral transcripts from cells infected with dl309, JM17-55R, dl521, or dl312. Samples were treated with DNase I to remove contaminating viral DNA, and no-RT controls from 96 hpi were run with each sample to ensure the efficiency of this process. All samples were normalized internally to GAPDH (A to E) and then to the levels of gene expression observed in dl312-infected cells (A to D), using the ΔΔCT method.
Levels of E3A expression during dl309 infection ranged from approximately 26,000-fold at 24 hpi to 1.2-million-fold at 48 hpi. Induction of E3A expression was much more modest during infection with 55R E1A-expressing viruses. At 24 hpi, E3A was induced only 2- and 7-fold by JM17-55R and dl521, respectively. Maximum E3A induction reached 23,000-fold at 144 hpi during dl521 infection and 2,900-fold at 120 hpi during JM17-55R infection. Again, both the kinetics and magnitude of E3A expression differed during infection with 55R E1A-expressing viruses compared to that with dl309, expressing wild-type E1A (Fig. 5B).
Induction of E4orf6/7 expression by dl309 infection was approximately 570-fold at 24 hpi, which is far less robust than that observed for E1B and E3A. Still, this level of induction was greater than those observed during JM17-55R and dl521 infections, which reached only 70- and 37-fold, respectively. Expression of E4orf6/7 peaked at 120 hpi with dl309, at over 5-million-fold induction. JM17-55R and dl521 also induced maximal expression at this time, albeit to far lower levels (4,500- and 10,000-fold, respectively) (Fig. 5C).
Finally, activation of the major late promoter (MLP) was determined by assessing the levels of hexon mRNA expression induced by each virus. During dl309 infection, hexon transcript levels increased progressively, from 46-fold above the level with dl312 at 24 hpi to a maximum of 298,000-fold above the level with dl312 at 120 hpi. For comparison, hexon was induced only 7-fold by JM17-55R at 24 hpi, and not at all by dl521. By 96 hpi, dl521 infection had reached its maximum degree of hexon expression, at 102,000-fold greater than that of dl312, while induction by JM17-55R reached a plateau at 2,100- and 1,200-fold at 120 and 144 hpi, respectively (Fig. 5D).
Despite the fact that dl312 lacks expression of all E1A isoforms, low levels of baseline viral transcripts could be detected in dl312-infected cells. In Fig. 5A to D, viral gene transcription has been normalized to that in samples infected with dl312 to account for baseline transcription levels. For clarity, we also show the relative level of viral gene transcription arising in dl312-infected cells at 144 hpi, normalized internally to GAPDH (Fig. 5E).
Taken together, these results show that 55R E1A is able to activate the expression of viral genes. This was true in the case of both dl521, which contains genomic E1A but splices only the 9S product, and JM17-55R, which carries 55R E1A cDNA in an E1A-deleted background (Fig. 1B). In each case, the levels of viral gene expression observed were distinct (and usually several log-fold lower at peak expression) from those of virus harboring wild-type E1A, as were the kinetics of gene expression. The ability of 55R E1A to activate viral genes is particularly interesting in light of the fact that 55R E1A does not contain any of the CRs present in the larger E1A isoforms. Further elucidation of the mechanism through which 55R E1A accomplishes this function will be of great interest.
55R E1A is sufficient to promote growth of HAdV in contact-inhibited IMR-90 fibroblasts.
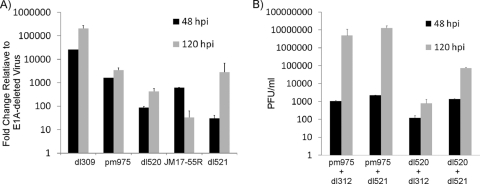
The observation that 55R E1A was able to activate expression of viral genes led us to investigate whether this E1A isoform also plays a role in promoting virus growth. Contact-inhibited IMR-90 cells were infected with dl309, pm975, dl520, JM17-55R, dl521, or dl312 at an MOI of 5. Supernatants were collected at 4, 48, and 120 hpi, and the yield of cell-free virus was assessed by plaque assay on HEK293 cells. In samples collected at 4 hpi, no more than 40 PFU/ml were ever present, and there was no significant difference between dl312 and the viruses expressing various E1A proteins (data not shown). Since growth was measured as fold changes relative to the growth of dl312 (E1A-deleted virus), this demonstrated that washing of cells 1 h after adsorption efficiently removed residual virus particles, that virus yields at later times were due to de novo virus replication, and that any growth advantage greater than that exhibited by dl312 could be attributed to the presence of the E1A isoform(s) expressed by the individual viruses.
As expected, dl309 grew best on the contact-inhibited IMR-90 fibroblasts, followed by pm975 (does not express 243R and 171R E1A). JM17-55R produced approximately 7-fold more virus than dl520 (does not express 289R and 217R E1A) at 48 hpi, while at 120 hpi this trend was reversed, with dl520 producing about 14-fold more virus than JM17-55R. dl521 grew to titers which closely resembled those reached by dl520 at both 48 and 120 hpi. Differences between dl521 and JM17-55R were likely due to delayed splicing of the 9S E1A transcript during dl521 infection, while it was expressed constitutively during JM17-55R infection. Most importantly, viruses expressing only 55R E1A consistently exhibited a 30- to 2,800-fold growth advantage compared to E1A-deleted virus, demonstrating that on its own, 55R E1A is sufficient to promote replication of HAdV in contact-inhibited IMR-90 cells (Fig. 6A). Therefore, not only can 55R E1A activate expression of viral genes, but it is also sufficient to promote viral replication.
Fig 6.
Replication of viruses expressing 55R E1A in contact-inhibited IMR-90 cells. (A) Contact-inhibited primary IMR-90 cells were infected with dl309, pm975, dl520, JM17-55R, dl521, or dl312 at an MOI of 5. Supernatants were collected at 4, 48, or 120 hpi, and cell-free virus titers were determined by serial dilution on HEK293 cells. (B) Contact-inhibited primary IMR-90 fibroblasts were infected with pm975 and dl312, pm975 and dl521, dl520 and dl312, or dl520 and dl521. Supernatants were collected at 4, 48, and 120 hpi, and the titers of cell-free virus were assayed as described for panel A.
Due to the genetic organization of genomic E1A, it was not possible to construct a virus that lacks expression of only 55R E1A without inducing mutations in the other E1A isoforms. Such a strategy runs the serious risk of confounding the interpretation of results gathered from this type of approach due to the large numbers of proteins with which E1A interacts. Nevertheless, it was important to evaluate the impact of 55R E1A on virus growth in the context of the two major E1A isoforms, 289R and 243R E1A. To accomplish this, we made use of a coinfection model whereby growth-arrested IMR-90 fibroblasts were infected with either pm975 or dl520 in combination with either dl521 or the E1A-deleted virus dl312. Cells were infected with each virus at an MOI of 5, for a total MOI of 10. In combination with a virus expressing 289R E1A (pm975), coinfection with dl521 only moderately improved virus yield compared to coinfection with dl312. However, coinfection of dl521 with a virus expressing 243R E1A (dl520) resulted in a 10-fold growth increase at 48 hpi relative to coinfection with dl312. This improved to a 100-fold increase by 120 hpi (Fig. 6B). It is important that both pm975 and dl520 can produce 55R E1A on their own, as a result of alternative splicing. Thus, these results suggest that in combination with 289R E1A, endogenous levels of 55R E1A are sufficient to maximize virus replication in contact-inhibited IMR-90 fibroblasts. However, in the context of 243R E1A, additional 55R E1A provided by coinfection with dl521 has a growth-promoting effect, which implies a unique mechanism for the replication-promoting phenotype exhibited by 55R E1A.
55R E1A interacts with the S8 component of the 19S regulatory proteasome.
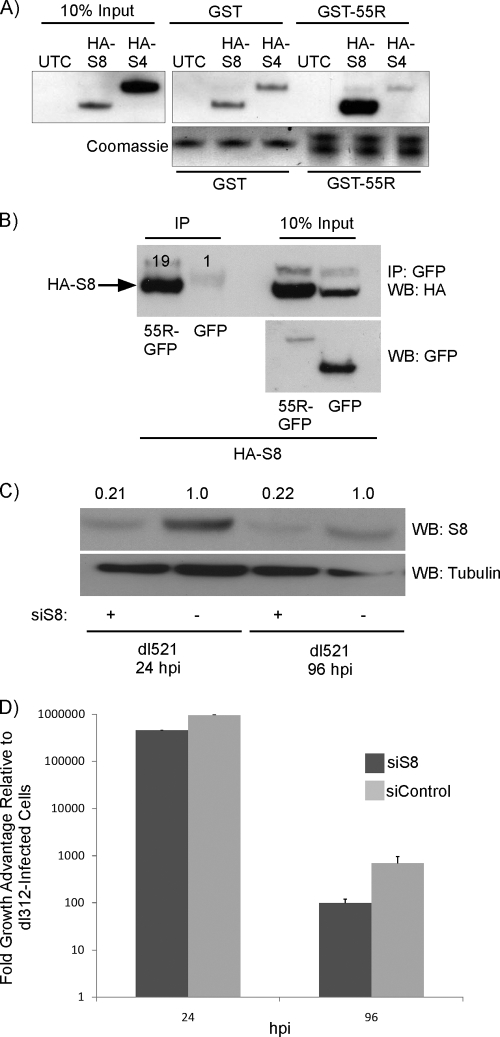
Interaction of the major E1A isoforms with the APIS complex has been demonstrated previously and is known to be important for the ability of E1A to enhance transcription of early genes (18). Binding of E1A to the S4 and S8 components of APIS was initially mapped to amino acids 4 to 25 of the E1A protein (29). Later studies showed that S8 could also be recruited by CR3 (18). In light of the fact that 55R E1A shares sequence identity with the larger E1A isoforms in its first 28 amino acids, we sought to determine whether it too could interact with components of APIS and whether this interaction was important for its ability to enhance virus replication.
To determine whether 55R E1A could interact with S4 and/or S8, A549 cells were transfected with a construct expressing either HA-S4 or HA-S8 or were mock transfected as a control. Lysates from these cells were incubated with GST-purified HAdV-2 55R E1A, which was pulled down with glutathione Sepharose beads. Interestingly, an interaction of GST-55R E1A with HA-S8, but not with HA-S4, was observed. Although there was evidence of some nonspecific binding of S8 to GST, binding of S8 to GST-55R E1A was 2.5 times more potent (Fig. 7A). To confirm that this interaction occurred in a more natural setting, we also cotransfected A549 cells with constructs expressing 55R E1A-GFP or GFP alone with constructs expressing HA-S8. Indeed, 55R E1A-GFP was able to pull down HA-S8, whereas GFP alone could not (Fig. 7B). In these assays, the quantities of input 55R E1A-GFP were much lower than those of GFP alone, likely due to the short half-life of the 55R E1A-GFP protein. Despite this, 55R E1A-GFP was able to pull down 19-fold more HA-S8 than GFP, providing strong support for the strength and specificity of this interaction.
Fig 7.
Interaction and consequences of 55R E1A binding to the APIS component S8. (A) Lysates were prepared from A549 cells transiently transfected with constructs expressing either HA-S8 or HA-S4. Lysates were incubated with GST-purified 55R E1A, and complexes were pulled down using glutathione Sepharose beads. The membrane was probed with anti-HA (3F10). (B) Lysates were prepared from A549 cells transiently transfected with constructs expressing HA-S8 and either 55R E1A-GFP or GFP alone. Complexes were immunoprecipitated using anti-GFP (Clontech) and protein A Sepharose beads. The membrane was probed with anti-HA (3F10) or anti-GFP (Clontech). (C and D) A549 cells were treated with scrambled siRNA or siRNA directed against S8. The cells were infected with either dl521 or dl312, and supernatants were collected at 48 hpi. (C) S8 levels were reduced by 79% at 24 hpi and by 78% at 96 hpi. (D) Virus titers were determined by serial dilution of supernatants on HEK293 cells.
To determine whether this interaction had functional consequences in the context of virus replication, endogenous S8 was knocked down in A549 cells by use of a validated siRNA directed against S8. We infected cells with either dl312 or dl521 and normalized dl521 growth to that observed using dl312 in cells treated with siRNA directed against S8 (siS8) or a scrambled siRNA control. This method of analysis allowed us to control for any nonspecific effects of S8 knockdown on virus growth and cellular health in general, thereby ensuring that the potential effects of S8 knockdown could be attributed directly to its interaction with 55R E1A. Knockdown of S8 was very effective at both 24 and 96 hpi, reaching 79% and 78% knockdown compared to control siRNA-treated cells, respectively (Fig. 7C). Interestingly, at 48 hpi, dl521 growth in cells treated with siS8 was reduced 2-fold compared to that in cells treated with control siRNA (Fig. 7D). The inhibitory effect of S8 knockdown on dl521 growth was amplified further by 96 hpi, at which time a 7-fold decrease in viral titer was observed in siS8-treated cells (Fig. 7D). This led us to conclude that the interaction of 55R E1A with S8 is functionally important for the ability of 55R E1A to promote virus replication. While the specific mechanism for the replication-promoting phenotype of the 55R E1A-S8 interaction will be the focus of future studies, it is possibly related to the ability of 55R E1A to activate viral gene expression, analogous to the case for the larger E1A isoforms.
DISCUSSION
Despite the discovery of the 9S E1A mRNA species over 30 years ago, the protein that it encodes, as well as the function of that protein, remained elusive. Early studies demonstrated that the 9S mRNA product is processed preferentially at late times postinfection and that this shift in splice site preference seems to require viral DNA replication, likely due to the shift in splicing factor usage following MLP activation and late transcript splicing (4, 25, 26, 32, 37). Unfortunately, the kinetics of 55R E1A protein expression could not be assessed at that time. To our knowledge, none of the E1A-directed Abs generated so far have been able to specifically detect the 55R E1A protein. This limitation has hampered efforts to biochemically and functionally characterize this novel E1A species.
To help address this problem, polyclonal rabbit Abs were generated against a peptide corresponding to amino acids 43 to 55 of the unique C-terminal region of the 55R E1A protein from HAdV-2. These Abs specifically recognized GST-purified 55R E1A from HAdV-2 but not the closely related HAdV-5 55R E1A protein or the equivalent but more distantly related 53R E1A protein encoded by HAdV-12 (Fig. 1C). The utility of these Abs was demonstrated in dot blot, Western blot, immunoprecipitation, and indirect immunofluorescence assays. The development of this reagent will aid in the further study of the HAdV-2 55R E1A protein, especially given the breadth of assays for which it is useful.
To dissect the functional role of 55R E1A during infection, it was important to first determine its expression kinetics and subcellular localization. Relative to the larger E1A isoforms, 55R E1A was expressed late during infection. During infection, 55R E1A displayed diffuse nuclear-cytoplasmic localization. Expression of tagged 55R E1A constructs in U2OS or HT1080 cells revealed that myc-tagged 55R E1A also exhibited primarily dual nuclear-cytoplasmic localization. Conversely, the EGFP-tagged constructs were largely excluded from the nucleus in the majority of cells (Fig. 4A and B). Since EGFP is over 4 times as large as 55R E1A, there is a high probability that the EGFP tag alters the natural localization of the protein. While 55R E1A does not contain a nuclear localization signal (NLS), the ability of 55R E1A to enter the nucleus may be explained by the fact that molecules of <40 kDa are able to pass relatively freely through the nuclear pore complex (34). At 6 kDa, 55R E1A falls well below this limit. The small size of the protein may also result in very rapid turnover. This is coupled with the fact that the second N-terminal residue of 55R E1A is an arginine, which is a destabilizing residue based on the N-end rule of protein degradation (31). In addition, the C-terminal domain is very rich in serine and proline residues, giving it PEST-like characteristics, which may also contribute to rapid turnover (20).
The localization of 55R E1A in the nucleus led us to investigate whether, akin to the larger E1A isoforms, 55R E1A had any role in activation of viral gene expression. No existing studies have directly examined the role of 55R E1A on viral replication. However, work examining the role of E1B 19K in regulating early gene expression hinted at some interesting properties of a virus carrying 9S cDNA in an E1A-deleted background. Namely, while E1B 19K repressed viral gene expression in HeLa cells infected with cDNA viruses expressing 289R and 243R E1A, it had a stimulatory effect on early gene expression in cells infected with a cDNA virus expressing 55R E1A (35, 36). While the particular reasons for this phenotype remain unclear, it led us to wonder whether 55R E1A could also activate viral gene expression during infection of contact-inhibited primary IMR-90 cells.
Indeed, our studies showed that viruses expressing only 55R E1A were sufficient to activate expression of both early and late viral genes beyond that observed during infection with an E1A-deleted virus. While the kinetics and magnitude of this activation varied from gene to gene, as expected, the maximal levels of expression induced were consistently lower than those exhibited by virus expressing wild-type E1A (Fig. 5). In the context of infection with wild-type virus, these effects were likely to be most pronounced at late times, when 55R E1A expression is at its peak. Since 55R E1A does not contain any of the CRs, the mechanism through which it is able to activate viral gene expression remains of great interest. This may be mediated in part through interactions with proteins known to bind within the first 28 amino acids of E1A, which are conserved in 55R E1A. In addition, it is possible that 55R E1A interacts with novel partners through its 27 unique C-terminal amino acids. This region is predicted to be unstructured and does not contain any easily recognizable domains that would help to predict binding partners or function. An in-depth, systematic characterization of this region will be the focus of future work.
While the 55R E1A protein was able to stimulate expression of viral genes in contact-inhibited IMR-90 cells, it did not guarantee that this was sufficient to allow the virus to replicate productively under these conditions. Although E1A-deleted viruses can undergo low-level replication in transformed cell lines, they replicate poorly unless high MOIs are used for infection (16). Therefore, we assessed the replication of HAdVs expressing various combinations of E1A proteins in contact-inhibited primary IMR-90 fibroblast cells infected at an MOI of 5. In all cases, replication was normalized to that observed during infection with dl312 (an E1A-null virus), such that any growth above this level could be attributed to a replication-promoting role for the respective E1A protein(s) carried by each virus. As expected, dl309 harboring wild-type genomic E1A replicated to the highest titers at both 48 and 120 hpi. This was followed by pm975, which expresses 289R E1A but not 243R E1A. dl520, which expresses 243R E1A but not 289R E1A, grew to titers approximately 2 and 3 log higher than those of dl312 at 48 and 120 hpi, respectively. Surprisingly, JM17-55R, carrying 9S cDNA in an E1A-deleted background, and dl521, which expresses 55R E1A, grew to equivalent, and in some cases higher, titers compared to those of dl520 (Fig. 6A). Again, this was unexpected given the lack of CRs present in 55R E1A and coupled with the observation that this protein does not have the potent transforming effects on cells seen with its larger counterparts (8). The differences observed between growth of JM17-55R and dl521 likely reflect both the expression level and kinetics of 55R E1A. While both viruses express 55R E1A under the control of the E1 promoter, it is expressed constitutively by JM17-55R and must be spliced from genomic E1A during infection with dl521.
During natural infection, 55R E1A is not expressed alone but in the context of the other E1A isoforms. We examined the effect of 55R E1A on virus replication in the presence of the larger E1A proteins. Coinfection experiments revealed that infection of cells with pm975 and dl521 did not enhance virus replication beyond that observed in cells infected with pm975 and dl312. This suggests that the endogenous levels of 55R E1A produced by pm975 are sufficient to maximize virus replication in combination with 289R E1A. However, coinfection of cells with dl520 and dl521 enhanced virus titers 1 and 2 log relative to those in cells infected with dl520 and dl312 at 48 and 120 hpi, respectively. These results indicate that in the context of 243R E1A, addition of exogenous 55R E1A can enhance virus replication (Fig. 6B). This observation is especially intriguing given that 243R E1A causes only low levels of viral gene transactivation on its own, while 289R E1A can activate viral gene expression to wild-type levels by itself (15, 38). Together, these observations lend even more strength to the evidence that 55R E1A directly activates expression of viral genes, which promotes productive virus replication.
Finally, it was determined that like 289R and 243R E1A, 55R E1A is able to bind S8, a regulatory component of the 26S proteasome and a member of the APIS complex. This is the first reported cellular binding partner of 55R E1A (Fig. 5A and B). The binding site for S8 was originally mapped to residues 4 to 25 of E1A (29). These residues are also present in the 55R E1A isoform. S4, another member of APIS, was also found to bind residues 4 to 25 of E1A. Notably, 55R E1A did not appear to bind this APIS subunit (29). The reason for this selectivity is unclear but may be a result of an inhibitory effect exerted by the unique C terminus of 55R E1A on the binding of S4. Alternatively, binding to S4 may require additional regions of the larger E1A proteins not present in 55R E1A.
We hypothesized that 55R E1A interaction with S8 might be required for virus replication. Knockdown of S8 reduced growth of dl521 2-fold by 24 hpi and 7-fold by 96 hpi in A549 cells (Fig. 7D). Since growth was normalized to that of dl312, this effect could be attributed fully to the effect of S8 knockdown on the function of 55R E1A. Therefore, in addition to identifying the first binding partner of 55R E1A, we have also determined that this interaction directly influences the ability of the 55R E1A protein to promote virus replication.
In conclusion, this is the first report to observe and functionally characterize 55R E1A since its discovery over 30 years ago. Further studies focused on unraveling the mechanism of 55R E1A-mediated viral gene transactivation and its growth-promoting properties will be important in understanding novel mechanisms controlling viral gene regulation. In addition, identification of putative binding partners of the unique C-terminal region of 55R E1A is likely to enhance our understanding of the interactions of HAdV with host cells and, more specifically, the E1A-mediated events that are important at late times postinfection.
ACKNOWLEDGMENTS
This work was supported by an operating grant from the Canadian Institutes of Health Research to J.S.M. and an NSERC operating grant to P.W. M.S.M. was supported by a Frederick Banting and Charles Best CIHR Canada Graduate Scholarship. S.D.B. was supported by a scholarship award from The Ontario HIV Treatment Network and a grant from the Canadian Institutes of Health Research (HIV/AIDS). M.J.C. was supported by graduate scholarships from the Canadian Institutes for Health Research (CIHR) Strategic Training Program. J.N.K. was supported in part by the Ontario Graduate Scholarship Program.
Footnotes
Published ahead of print 1 February 2012
REFERENCES
- 1. Abou El Hassan MA, Braam SR, Kruyt FA. 2006. A real-time RT-PCR assay for the quantitative determination of adenoviral gene expression in tumor cells. J. Virol. Methods 133:53–61 [DOI] [PubMed] [Google Scholar]
- 2. Berk AJ. 2007. Adenoviridae, p 2355–2394 In Knipe DM, et al. (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 3. Berk AJ, Sharp PA. 1978. Structure of the adenovirus 2 early mRNAs. Cell 14:695–711 [DOI] [PubMed] [Google Scholar]
- 4. Chow LT, Broker TR, Lewis JB. 1979. Complex splicing patterns of RNAs from the early regions of adenovirus-2. J. Mol. Biol. 134:265–303 [DOI] [PubMed] [Google Scholar]
- 5. Dijkema R, et al. 1980. Gene organization of the transforming region of weakly oncogenic adenovirus type 7: the E1a region. Gene 12:287–299 [DOI] [PubMed] [Google Scholar]
- 6. Ferguson B, et al. 1985. E1A 13S and 12S mRNA products made in Escherichia coli both function as nucleus-localized transcription activators but do not directly bind DNA. Mol. Cell. Biol. 5:2653–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Grand RJ, et al. 1999. Adenovirus early region 1A protein binds to mammalian SUG1—a regulatory component of the proteasome. Oncogene 18:449–458 [DOI] [PubMed] [Google Scholar]
- 8. Haley KP, Overhauser J, Babiss LE, Ginsberg HS, Jones NC. 1984. Transformation properties of type 5 adenovirus mutants that differentially express the E1A gene products. Proc. Natl. Acad. Sci. U. S. A. 81:5734–5738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Harlow E, Franza BR, Jr, Schley C. 1985. Monoclonal antibodies specific for adenovirus early region 1A proteins: extensive heterogeneity in early region 1A products. J. Virol. 55:533–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hilleman MR, Werner JH. 1954. Recovery of new agent from patients with acute respiratory illness. Proc. Soc. Exp. Biol. Med. 85:183–188 [DOI] [PubMed] [Google Scholar]
- 11. Hua S, Sun Z. 2001. Support vector machine approach for protein subcellular localization prediction. Bioinformatics 17:721–728 [DOI] [PubMed] [Google Scholar]
- 12. Jones N, Shenk T. 1979. An adenovirus type 5 early gene function regulates expression of other early viral genes. Proc. Natl. Acad. Sci. U. S. A. 76:3665–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jothikumar N, et al. 2005. Quantitative real-time PCR assays for detection of human adenoviruses and identification of serotypes 40 and 41. Appl. Environ. Microbiol. 71:3131–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lyons RH, Ferguson BQ, Rosenberg M. 1987. Pentapeptide nuclear localization signal in adenovirus E1a. Mol. Cell. Biol. 7:2451–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Montell C, Fisher EF, Caruthers MH, Berk AJ. 1982. Resolving the functions of overlapping viral genes by site-specific mutagenesis at a mRNA splice site. Nature 295:380–384 [DOI] [PubMed] [Google Scholar]
- 16. Nevins JR. 1981. Mechanism of activation of early viral transcription by the adenovirus E1A gene product. Cell 26:213–220 [DOI] [PubMed] [Google Scholar]
- 17. Perricaudet M, Akusjarvi G, Virtanen A, Pettersson U. 1979. Structure of two spliced mRNAs from the transforming region of human subgroup C adenoviruses. Nature 281:694–696 [DOI] [PubMed] [Google Scholar]
- 18. Rasti M, et al. 2006. Roles for APIS and the 20S proteasome in adenovirus E1A-dependent transcription. EMBO J. 25:2710–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Roberts BE, et al. 1985. Individual adenovirus type 5 early region 1A gene products elicit distinct alterations of cellular morphology and gene expression. J. Virol. 56:404–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rogers S, Wells R, Rechsteiner M. 1986. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science 234:364–368 [DOI] [PubMed] [Google Scholar]
- 21. Rowe WP, Huebner RJ, Gilmore LK, Parrott RH, Ward TG. 1953. Isolation of a cytopathogenic agent from human adenoids undergoing spontaneous degeneration in tissue culture. Proc. Soc. Exp. Biol. Med. 84:570–573 [DOI] [PubMed] [Google Scholar]
- 22. Shen HB, Chou KC. 2010. Virus-mPLoc: a fusion classifier for viral protein subcellular location prediction by incorporating multiple sites. J. Biomol. Struct. Dyn. 28:175–186 [DOI] [PubMed] [Google Scholar]
- 23. Shen HB, Chou KC. 2007. Virus-PLoc: a fusion classifier for predicting the subcellular localization of viral proteins within host and virus-infected cells. Biopolymers 85:233–240 [DOI] [PubMed] [Google Scholar]
- 24. Shenk T, Jones N, Colby W, Fowlkes D. 1980. Functional analysis of adenovirus-5 host-range deletion mutants defective for transformation of rat embryo cells. Cold Spring Harb. Symp. Quant. Biol. 44:367–375 [DOI] [PubMed] [Google Scholar]
- 25. Spector DJ, Halbert DN, Raskas HJ. 1980. Regulation of integrated adenovirus sequences during adenovirus infection of transformed cells. J. Virol. 36:860–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Spector DJ, McGrogan M, Raskas HJ. 1978. Regulation of the appearance of cytoplasmic RNAs from region 1 of the adenovirus 2 genome. J. Mol. Biol. 126:395–414 [DOI] [PubMed] [Google Scholar]
- 27. Stephens C, Harlow E. 1987. Differential splicing yields novel adenovirus 5 E1A mRNAs that encode 30 kD and 35 kD proteins. EMBO J. 6:2027–2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Trentin JJ, Yabe Y, Taylor G. 1962. The quest for human cancer viruses. Science 137:835–841 [DOI] [PubMed] [Google Scholar]
- 29. Turnell AS, et al. 2000. Regulation of the 26S proteasome by adenovirus E1A. EMBO J. 19:4759–4773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ulfendahl PJ, et al. 1987. A novel adenovirus-2 E1A mRNA encoding a protein with transcription activation properties. EMBO J. 6:2037–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Varshavsky A. 1997. The N-end rule pathway of protein degradation. Genes Cells 2:13–28 [DOI] [PubMed] [Google Scholar]
- 32. Virtanen A, Pettersson U. 1983. The molecular structure of the 9S mRNA from early region 1A of adenovirus serotype 2. J. Mol. Biol. 165:496–499 [DOI] [PubMed] [Google Scholar]
- 33. Wang W, Chevray PM, Nathans D. 1996. Mammalian Sug1 and c-Fos in the nuclear 26S proteasome. Proc. Natl. Acad. Sci. U. S. A. 93:8236–8240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wente SR, Rout MP. 2010. The nuclear pore complex and nuclear transport. Cold Spring Harb. Perspect. Biol. 2:a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. White E, Denton A, Stillman B. 1988. Role of the adenovirus E1B 19,000-dalton tumor antigen in regulating early gene expression. J. Virol. 62:3445–3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. White E, Stillman B. 1987. Expression of adenovirus E1B mutant phenotypes is dependent on the host cell and on synthesis of E1A proteins. J. Virol. 61:426–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wilson MC, Darnell JE., Jr 1981. Control of messenger RNA concentration by differential cytoplasmic half-life. Adenovirus messenger RNAs from transcription units 1A and 1B. J. Mol. Biol. 148:231–251 [DOI] [PubMed] [Google Scholar]
- 38. Winberg G, Shenk T. 1984. Dissection of overlapping functions within the adenovirus type 5 E1A gene. EMBO J. 3:1907–1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wold WSM, Tollefson AE. 2007. Adenovirus methods and protocols: adenoviruses, Ad vectors, quantitation and animal models. Humana Press, Totowa, NJ [Google Scholar]