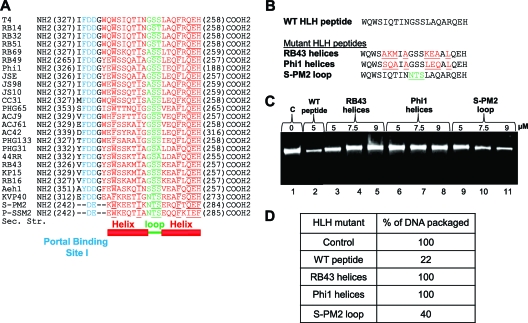
Fig 7.
Dissecting the functional roles of residues in the helix and loop regions of the HLH motif. (A) Sequence alignment of the HLH motifs of T4 family large terminases. The helices are shown in red and the loop in green. Highly conserved amino acids (>90% identity) are underlined. The portal binding site I (Fig. 4) is highlighted in cyan. (B) Sequences of HLH swap mutants. The T4 (WT) sequence is shown in black. Mutations introduced in the helix region (red) correspond to sequences of phages RB43 and Phi1. Mutations introduced in the loop region (green) correspond to the sequence of phage S-PM2. The F348A mutation was incorporated into all the constructs because it enhanced the solubility of the E. coli-expressed Soc-fused recombinant peptides. (C) Effects of swap mutant Soc-HLH peptides on DNA packaging. DNA packaging assays were performed according to the procedure described in Materials and Methods. Proheads (4 × 109 particles) were incubated with increasing concentrations of the mutant peptides (5, 7.5, or 9 μM) for 15 min. The agarose gel shows the amount of DNA packaged in the absence of peptide (lane 1), in the presence of WT peptide (lane 2) or increasing amounts of mutant peptides (lanes 3 to 11). (D) Relative amounts of DNA packaged in the reactions in panel C.