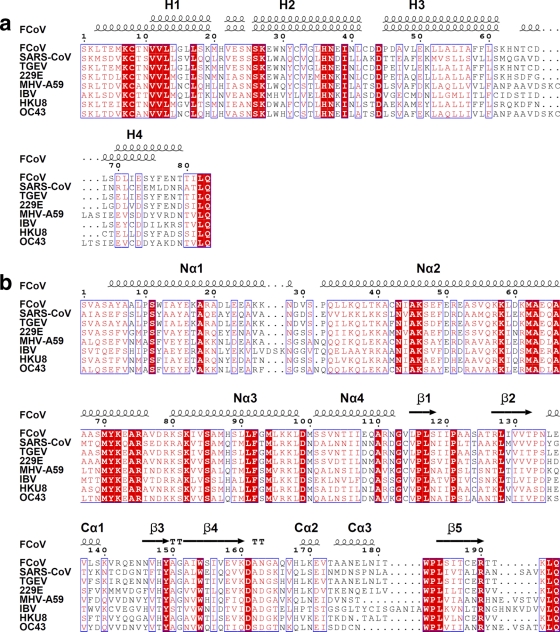
Fig 3.
Sequence alignment of FCoV Nsp7 (a) and Nsp8 (b) with homologues from other coronaviruses. The alignments were achieved by using CLUSTAL W2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/), and colored figures were generated by ESPript2.2 (http://espript.ibcp.fr/ESPript/ESPript/). Residues labeled in red are conserved to >70%, and residues boxed in red are completely identical. Secondary structures of FCoV Nsp7 (above, Nsp7II; below, Nsp7I) and Nsp8 are indicated at the top of the alignments. All of the sequences aligned here are from the National Center for Biotechnology Information (NCBI) database. Abbreviations and accession numbers: feline infectious peritonitis virus (FCoV), DQ010921.1; SARS-CoV, NP_828865 (Nsp7) and NP_828866 (Nsp8); transmissible gastroenteritis coronavirus (TGEV), NP_840005.1 (Nsp7) and NP_840006.1 (Nsp8); human coronavirus 229E (229E), NP_835348.1 (Nsp7) and NP_835349.1 (Nsp8); mouse hepatitis virus strain A59 (MHV-A59), NP_740612.1 (Nsp7) and NP_740613.1 (Nsp8); infectious bronchitis virus (IBV), NP_740625.1 (Nsp7) and NP_740626.1 (Nsp7); bat coronavirus HKU8, YP_001718610.1; and human coronavirus OC43 (OC43), NP_937947.