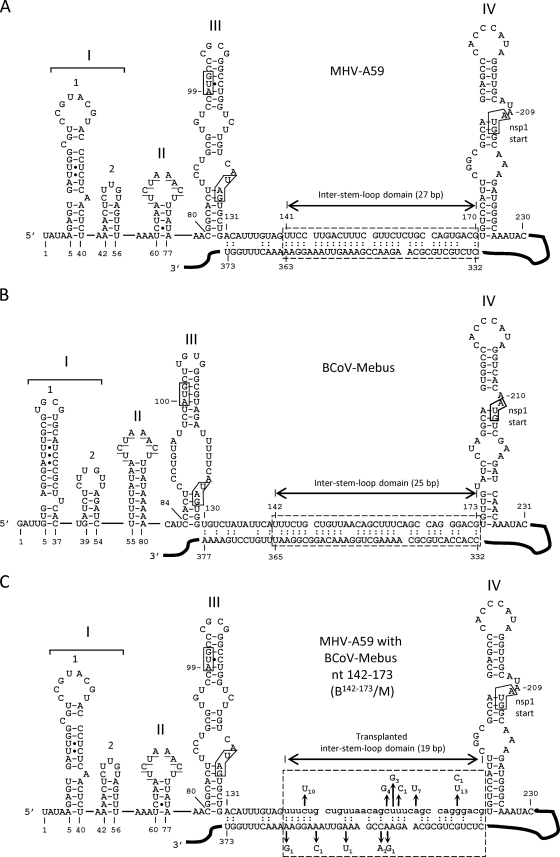
Fig 1.
5′ UTRs of MHV-A59 and BCoV-Mebus showing the intra-5′-UTR stem-loops, the inter-stem-loop domain, and a potential long-range RNA-RNA interaction between the 5′ UTR and the Nsp1 coding region. (A) 5′ UTR of MHV showing the Mfold-predicted long-range RNA-RNA interaction (hatched box). (B) 5′ UTR of BCoV showing the Mfold-predicted long-range RNA-RNA interaction (hatched box). (C) 5′ UTR of MHV in which the 32-nt BCoV inter-stem-loop domain (lowercase) replaces the 30-nt MHV counterpart. The hatched box identifies a potential long-range interaction and shows a summary of the potential suppressor mutations in 17 plaque-purified clones described in Table 1. The subscript for each mutation indicates its incidence among the 17 isolates. Note that without suppressor mutations, the number of base pairs within the window is 19, not 27 (as in MHV) or 25 (as in BCoV). Refolding enables the number of base pairs to increase as a function of suppressor mutations (see the text). In all panels, the underlined bases in SLII identify the transcription-regulatory sequence (UCUAAAC), and boxes in SLIII identify the start and stop codons for a short upstream open reading frame. The box in SLIV identifies the start codon for the Nsp1 coding region. Filled circles in SL1, SLII, and SLIII represent non-Watson-and-Crick base pairings, as determined by nuclear magnetic resonance studies (33, 36).