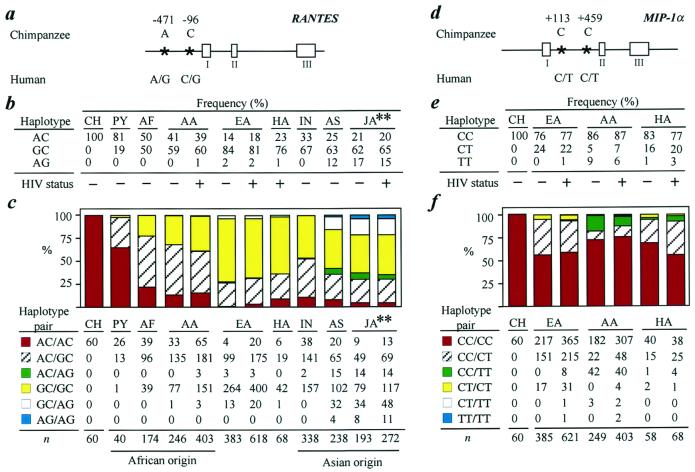
Figure 1.
(a–c) RANTES and (d–f) MIP-1α SNPs, haplotypes, and haplotype pairs. (a and d) Schematic illustration of the genomic organization and SNPs (*) in human RANTES and MIP-1α. Open boxes represent exons. The sequence at the position corresponding to the human SNPs in chimpanzees (ancestral state) is also shown. The designation of each haplotype is based on the 5′ > 3′ arrangement of the SNPs. Thus, for example, the haplotype that contains RANTES −471A and −96C is designated as the RANTES AC haplotype. (b and e) Frequencies of the RANTES and MIP-1α haplotypes in chimpanzee (CH) and control blood donors (−) and infected (+) worldwide populations, including European (EA), African (AA), and Hispanic Americans (HA). PY, pygmy; AF, non-Pygmy Africans; IN, Asian Indians; AS, non-Indian Asians; JA, Japanese. The ancestral haplotype for each gene is listed first in panels b and e. (c and f): (Upper) The frequency (%) of the RANTES and MIP-1α haplotype pairs in the different populations. (Lower) The total number of individuals with a given haplotype pair is shown. The color codes shown adjacent to each haplotype pair listed in the Lower correspond to the colored bars shown in the Upper. The ancestral haplotype pair for each gene is in red. **, data listed under the column heading designated as JA is derived from the study by Liu et al. (27), and shows the frequency of the RANTES haplotype (b) and haplotype pairs (c) in Japanese HIV− and HIV+ individuals; the data for the HIV+ and HIV− Japanese hemophiliacs and nonhemophiliacs was combined. The codes shown for the HIV status in b and e are also applicable for the data shown in c and f, respectively.