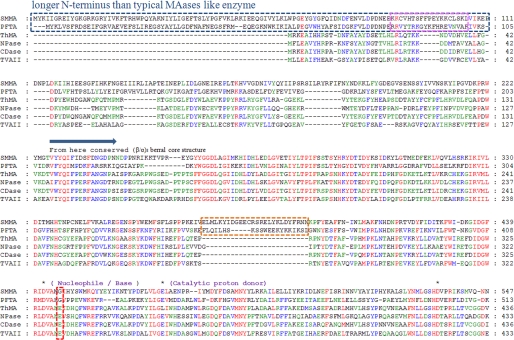
FIGURE 2.
Sequence alignment for SMMA, PFTA, ThMA, NPase, TVAII, and CDase. The sequences of six CD-hydrolyzing enzymes, two hyperthermophilic archaic maltogenic amylases, and four conventional bacterial CD-hydrolyzing enzymes were aligned using ClustalW (27). The novel SMMA N′ domain is indicated with a blue dotted line. The helix-loop-helix regions observed in the structure are indicated with orange dotted lines. The β-7-loop 7-β-8 region protruding from the N′ domain is indicated with a pink dotted line. The three conserved catalytic residues in the GH13 family are indicated with black asterisks. Red, blue, and green highlighting indicates the residues that are conserved in 100, 75, and 50% of the proteins, respectively. The accession numbers for these sequences are ABN69720.1 (SMMA), AAL82063.1 (PFTA from Pyrococcus furiosus), AAC15072.1 (ThMA from Thermus sp. strain IM6501), AAA22622.1 (NPase from B. stearothermophilus), BAA02473.1 (TVAII), and AAA92925.1 (CDase from alkalophilic Bacillus sp. I-5).