Background: Extra-large G proteins, XLGs, are nuclear GTP-binding proteins with both Gα-like and novel domains.
Results: Unlike general G proteins, XLG proteins are Ca2+-dependent GTPases, and XLG2 interacts with the nuclear protein RTV1 (related to vernalization 1).
Conclusion: GTP-bound XLG2 promotes RTV1-chromatin interaction, leading to activation of floral integrator genes.
Significance: XLGs unusually integrate Ca2+-based and G protein-based cellular pathways and control flowering.
Keywords: Arabidopsis thaliana; Chromatin Immunoprecipitation (ChiP); Heterotrimeric G Proteins; Plant Biochemistry; Transcription Regulation; Extra-large G Proteins; Flowering; GPA1, GTPase, RTV1, XLG
Abstract
Heterotrimeric G proteins, consisting of Gα, Gβ, and Gγ subunits, play important roles in plant development and cell signaling. In Arabidopsis, in addition to one prototypical G protein α subunit, GPA1, there are three extra-large G proteins, XLG1, XLG2, and XLG3, of largely unknown function. Each extra-large G (XLG) protein has a C-terminal Gα-like region and a ∼400 amino acid N-terminal extension. Here we show that the three XLG proteins specifically bind and hydrolyze GTP, despite the fact that these plant-specific proteins lack key conserved amino acid residues important for GTP binding and hydrolysis of GTP in mammalian Gα proteins. Moreover, unlike other known Gα proteins, these activities require Ca2+ instead of Mg2+ as a cofactor. Yeast two-hybrid library screening and in vitro protein pull-down assays revealed that XLG2 interacts with the nuclear protein RTV1 (related to vernalization 1). Electrophoretic mobility shift assays show that RTV1 binds to DNA in vitro in a non-sequence-specific manner and that GTP-bound XLG2 promotes the DNA binding activity of RTV1. Overexpression of RTV1 results in early flowering. Combined overexpression of XLG2 and RTV1 enhances this early flowering phenotype and elevates expression of the floral pathway integrator genes, FT and SOC1, but does not repress expression of the floral repressor, FLC. Chromatin immunoprecipitation assays show that XLG2 increases RTV1 binding to FT and SOC1 promoters. Thus, a Ca2+-dependent G protein, XLG2, promotes RTV1 DNA binding activity for a subset of floral integrator genes and contributes to floral transition.
Introduction
Signaling via heterotrimeric G proteins, composed of Gα, Gβ, and Gγ subunits, is highly conserved in eukaryotes (1–4). Classically, G protein-mediated signaling is initiated by ligand binding to a cell surface G protein-coupled receptor, which has seven transmembrane domains. Ligand binding activates receptor-mediated GDP/GTP exchange on the Gα subunit. Resultant GTP binding causes the dissociation of Gα from the Gβγ dimer, and the activated Gα and Gβγ interact with downstream target proteins, leading to numerous cellular responses (5–7). Although mammals have 800–1000 protein-coupled receptors, 23 Gα, 5 Gβ, and 12 Gγ subunits (8), plants have a greatly reduced number of heterotrimeric G protein signaling elements (9–11). For instance, the Arabidopsis genome encodes only one Gα subunit (GPA1), one Gβ subunit (AGB1), three Gγ subunits (AGG1, AGG2, and AGG3), and 1–2 dozen candidate G protein-coupled receptors (12–18). However, despite the small number of heterotrimeric G protein signaling elements, heterotrimeric G proteins play important roles in many physiological and developmental processes in plants (3, 10, 19).
Besides the canonical Gα subunit gene, GPA1, there are three genes in Arabidopsis that encode unusual extra-large GTP-binding proteins, known as XLGs (20, 21). All XLG proteins have C-terminal Gα domains that are homologous to GPA1 and other Gα subunits. The unique N-terminal regions of XLGs consist of ∼400 amino acid sequences that are highly conserved among the XLG proteins. XLG proteins appear to be unique to the plant kingdom and are not homologous to the XLαs of mammals (21). Despite variations in conserved amino acid residues required for GTP binding compared with canonical Gα proteins, recombinant XLG1 protein was previously shown to bind GTP, which is a functional characteristic of Gα proteins (20). Loss-of-function analyses showed redundancy in XLG function; dark-grown xlg1-1 xlg2-1 xlg3-1 triple mutant plants show an increased primary root length and altered sensitivity in root growth in response to sugar, abscisic acid, osmotic stress, and ethylene. In addition, XLG3 is specifically involved in the regulation of root waving and skewing (21, 22), and XLG2 interacts with the sole Arabidopsis canonical Gβ subunit, AGB1, and is involved in pathogen resistance (23). Expression analyses of XLG genes using RT-PCR and a reporter gene assay show that all three XLGs are widely expressed throughout the plant, suggesting that XLGs may have additional functions in plant development and signaling beyond those already reported (21).
In Arabidopsis, flowering time is controlled by multiple endogenous and environmental cues, including photoperiod, temperature, and gibberellins. Floral regulatory cues are integrated by a group of genes, including FT (flowering locus T), SOC1/AGL20 (suppressor of overexpression of constans 1), AGL24 (agamous-like 24), and LFY (leafy). These genes are often referred to as “floral integrators” because most known flowering pathways eventually regulate expression of these flowering genes (24–27). Studies in Arabidopsis and rice have shown that the floral integrator FT is a major component of florigen (28, 29). FT together with FD (flowering locus D) activates SOC1 in the meristem (30). Floral integrators, in turn, activate floral meristem identity genes, including AP1 (apetala 1) and FUL (fruitful) (30, 31). Thus, there is an intricate regulatory network among floral integrators and floral meristem identity genes to initiate and maintain floral transition in response to endogenous and environmental cues (24, 25, 27, 30, 31).
Winter annual strains of Arabidopsis contain functional FRI (frigida) and FLC (flowering locus C) genes and flower extremely late unless plants are exposed to a prolonged period of cold (i.e. vernalization). FLC acts as a potent floral repressor by the direct suppression of floral regulators FT and SOC1 (32, 33). Vernalization results in stable repression of FLC to allow rapid flowering (34). Similar to winter annual strains of Arabidopsis, a group of late flowering mutants flower late in an FLC-dependent manner (35). These mutants are collectively referred as the autonomous pathway mutants, and vernalization results in rapid flowering in the autonomous mutants as well (35). Vernalization results in an increase in two repressive histone modifications of FLC chromatin: methylation of histone H3 Lys-9 and histone H3 Lys-27 (34, 36). These vernalization-induced histone modifications are mediated by a number of proteins, including VRN2 (vernalization 2), VRN1 (vernalization 1), and VIN3 (vernalization-insensitive 3) (34, 36). In this study, we describe the biochemical characterization of the XLG2 protein and address its function in regulation of RTV1 (related to vernalization 1) and in floral transition, thereby elucidating a role of this unusual G protein in the control of flowering time.
EXPERIMENTAL PROCEDURES
Plant Materials and Vernalization Treatment
Seeds were germinated on 0.8% agar plates with half-strength Murashige and Skoog salt, 0.8% sucrose, stratified at 4 °C for 2 days, and grown for 10 days in long day conditions (16 h of light at 22 °C, and 8 h of dark at 22 °C) or short day conditions (8 h of light at 22 °C, 16 h of dark at 22 °C), and then plants were transplanted to soil or harvested for RNA isolation. Light intensity for LD2 and SD conditions was ∼70 μmol m−2 s−1. For the vernalization treatment, seeds were germinated on agar plates for 5 days in SD conditions, and vernalized for 5 weeks at 4 °C in SD (37), and then plants were harvested for RNA isolation, or plants were transplanted to soil and transferred to growth chambers (21 ± 2 °C) under LD or SD conditions for flowering time tests.
Protein Expression in Escherichia coli and Generation of XLG2(T475N) Mutant Construct
For expression in E. coli, the coding regions of the XLG genes and GPA1 cDNA were subcloned into the pET28a-modified expression vector (Novagen) which results in production of fusion proteins with N-terminal His6 and thioredoxin (TRX) tags. We fused XLG proteins with TRX to increase solubility of the recombinant proteins. The coding region of RTV1 and each XLG was also subcloned into the pGEX-2T vector, which produces GST fusion proteins. XLG C-terminal regions (amino acids 486–888 for XLG1, 461–862 for XLG2, and 433–849 for XLG3) were also subcloned into the same vector. All constructs were transformed into the E. coli BL21(DE3)pLysS strain. Transformed cells were inoculated into 10 ml of LB medium containing 25 μg/ml kanamycin for pET28a and 50 μg/ml ampicilin for pGEX-2T. The cultures were grown at 30 °C overnight and then poured into 500 ml of fresh LB medium. When an A600 of 0.8 was reached, isopropyl β-d-thiogalactopyranoside was added to the cultures to a final concentration of 0.9 mm, and the incubation was continued for another 4 h at 30 °C for induction. His-tagged fusion proteins were purified using Ni2+-nitrilotriacetic acid-agarose column chromatography (Qiagen) (supplemental Fig. 1, a and b), and GST fusion proteins were purified using glutathione-agarose (Sigma) according to each manufacturer's instructions. Empty vectors were transformed into the E. coli BL21 (DE3) pLysS strain and expressed for purification of His6 TRX or GST. To purify XLG proteins lacking any tag, GST was cleaved by thrombin (Sigma) from GST fusion XLG proteins. Mutant (T475N) forms of full-length XLG2 and XLG2C were constructed by PCR using the following primers: 5′- GGTGGCGCCAATACAATTTACAAA-3′ and 5′-TTTGTAAATTGTATTGGCGCCACC-3′.
GTP Binding Assay
The [35S]GTPγS binding activity of full length XLGs, C-terminal regions of the XLGs, GPA1, and TRX protein was measured by the rapid filtration method (38, 39). A 100 nm concentration of each purified recombinant proteins was incubated at 30 °C in 200 μl of 50 mm Tris-Cl buffer, pH 8.0, containing 1 mm EDTA, 1 mm dithiothreitol (DTT), 1 mm NaN3, 0.2 μm [35S]GTPγS, and 10 mm MgCl2 or CaCl2. The reaction was stopped by the addition of 2 ml of ice-cold buffer (20 mm Tris-Cl, pH 8.0, 100 mm NaCl, and 25 mm MgCl2). Aliquots (20 μl) were taken at the indicated times and vacuum-filtered onto nitrocellulose membranes (Millipore, Billerica, MA), which had been rinsed with 3 ml of ice-cold buffer, and membrane-bound material was washed rapidly three times with 3 ml of ice-cold buffer. The washed membranes were dried and quantified for radioactivity by liquid scintillation counting.
GTPase Assay
GTP hydrolysis assays were performed as previously described (40) using PEI-cellulose TLC plates. Briefly, the reaction was performed at 30 °C in 200 μl of buffer containing 50 mm Tris-Cl, pH 8.0, 1 mm EDTA, 1 mm dithiothreitol, 1 mm NaN3, 20 pmol of [α-32P]GTP, 100 nm full-length or C-terminal XLG protein or 100 nm of GPA1 protein, and 10 mm MgCl2, MnCl2 or CaCl2. Aliquots of 10 μl were sampled at appropriate time intervals, and 10 μl of 0.5 m EDTA (pH 8.0) was added to the aliquots to stop the reaction. Samples (2 μl) were spotted onto PEI-cellulose TLC plates, which were then developed in 0.5 m KH2PO4, pH 3.4, solution. After drying, the plates were exposed to a storage phosphor screen (Amersham Biosciences) and then scanned by a PhosphorImager (GE Healthcare).
Yeast Two-hybrid Screening
Yeast two-hybrid screening was carried out according to the manufacturer's protocol (Clontech). Briefly, the bait plasmid, pBUTE-XLG2, and total cDNAs of Arabidopsis obtained from ABRC (catalogue number CD4-10) in which the cDNAs were fused to the pACT-AD were used sequentially to transform Saccharomyces cerevisiae strain pJG69-4A. The resulting transformants were then plated on synthetic complete (SC) medium lacking uracil (Ura), leucine (Leu), and histidine (His) (SC−, Ura−, Leu−, and His−) and/or SC− Ura− and Leu−, including 3 mm 3-aminotriazole. Histidine-positive colonies were further tested for β-galactosidase (LacZ) activity according to the manufacturer's protocol (Clontech). The clones of cDNAs encoding candidate XLG2 interacting proteins were selected and isolated based on their growth as blue colonies in histidine-deficient medium. The cDNAs were isolated and sequenced. The DNA sequences were then subjected to BLAST analysis using the GenBankTM database.
In Vitro Protein Pull-down Assay
Purified GST or GST-RTV1 was incubated for 2 h at 4 °C with 40 μl of glutathione-Sepharose in PBST (PBS containing 1% Triton X-100). After three washes, the beads were incubated with His-XLG2C in 0.5 ml of PBST for 3 h on a rotary shaker at 4 °C. The beads were then washed five times with PBST, boiled with SDS sample buffer, and subjected to 12% SDS-PAGE. Western blot of the resolved proteins was carried out using anti-His (Qiagen catalog no. 34660) and anti-GST (Novagen catalog no. 71097) monoclonal antibodies.
Transient Expression Proteins Fused with Green Fluorescent Protein
RTV1 cDNA was fused upstream of the GFP cDNA under the control of the CaMV 35S promoter (pUC::GFP) (41). The construct (RTV1-GFP) was co-transformed into Arabidopsis protoplasts along with the red fluorescent protein cDNA fused to a nuclear localization signal peptide (NLS-RFP (42)). Transient expression of GFP- and RFP-fused constructs in Arabidopsis protoplasts was performed according to the method described by Heo et al. (40). Briefly, recombinant plasmids were introduced by polyethylene glycol (PEG)-mediated transformation into Arabidopsis protoplasts that had been prepared from leaf tissues. Expression of the fusion constructs was monitored after transformation and observed by fluorescence microscopy (Olympus AX70 TR, Olympus).
Production of Transgenic Arabidopsis Lines
For generation of PRTV1::GUS lines (Col background), the RTV1 promoter (0.91 kb, which is the complete intergenic region) was amplified and subcloned into the pENTR directional TOPO vector (Invitrogen) followed by LR reaction (Invitrogen) into the destination vector pHGWFS7 (43) using LR enzyme (Invitrogen). GUS activity was revealed by incubation in 100 mm NaPO4 (pH 7.2), 2.5 mm 5-bromo-4-chloro-3-indoyl-β-d-glucuronide. Plant tissue was incubated at 37 °C for 24 h. After staining, chlorophyll was cleared from the samples by dehydration using 70% ethanol.
To generate 35S-RTV1 lines, RTV1 cDNA was ligated to the pH7GWIWG2 vector, and the construct was transformed into wild-type plants (Col-0) via Agrobacterium-mediated transformation (44). For 35S-XLG2 RTV1, XLG2 cDNA was ligated to pGWB11, and the resulting 35S-XLG2 construct was transformed into the T1 generation of 35S-RTV1(13) plants. Transgenic lines were selected on selective antibiotic marker-containing medium and were confirmed by genotyping PCR. To generate 35S-RTV1-GFP ft-1 lines, the 35S-RTV1-GFP construct was transformed into ft mutant plants by floral dipping, and then T1 generation plants were selected by selective antibiotic-containing medium, followed by flowering time determination.
Electrophoretic Mobility Shift Assay (EMSA)
EMSAs were performed as described by Despres et al. (45). DNA fragments of FLC, SOC1, FT, VRN2, and DFR promoters 400 bp upstream of their start codons were generated by PCR amplification and confirmed by sequencing. 100 ng of double-stranded synthetic nucleotides were radiolabeled using T4 polynucleotide kinase. EMSA reactions were performed at room temperature for 30 min in 15 μl of EMSA buffer (20 mm Hepes-KOH, pH 7.9, 100 mm KCl, 2 mm DTT, and 20% glycerol) containing 1 μl (20,000 cpm) of the labeled nucleotides and 0–2 μg of RTV1. In addition, 100 nm XLG2C, 10 mm Ca2+, and 1 mm GTPγS were added with the buffer in some reactions. EMSA reactions were loaded onto a 5% Tris-borate EDTA-polyacrylamide gel and run at 8 V/cm for 90 min. After electrophoresis, the gel was dried and subjected to autoradiography at −80 °C using Eastman Kodak Co. x-ray film.
Chromatin Immunoprecipitation (ChIP) Assays and Immunoprecipitation
ChIP assays were carried out (Johnson and Bresnick (46)) as described by Heo et al. (47), following the protocol of Gendrel et al. (48). Chromatin proteins and DNA were cross-linked in 10-day-old Col, 35S-RTV1(13) and 35S-XLG2(1) RTV1 seedlings by formaldehyde fixation. Chromatin was isolated using the same extraction buffers as in Heo and Sung (47) and Gendrel et al. (48). Chromatin size postsonication was 500–800 bp, as confirmed by agarose gel electrophoresis (not shown). Chromatin immunoprecipitation was performed using anti-FLAG (Sigma; catalog no. F1804) or anti-GFP (Molecular Probes; catalog no. A11122) antibodies. Cross-links were reversed by incubation at 65 °C for 6 h, and DNA was purified with QIAquick spin columns (Qiagen) and eluted in 50 μl of Tris-HCl EDTA (pH 8.0). PCR was used to amplify fragments of the FT, SOC1, and FLC promoters as used in EMSAs. The relative -fold increase of retrieved DNAs was calculated as follows: first, relative enrichment of each fragment was calculated by comparison with the input (chromatin sample before immunoprecipitation). Second, the background levels of retrieved DNAs were calculated by measuring ACTIN2 enrichment compared with input. Third, relative levels of each retrieved fragment were calculated using the formula, CT = CT(each gene) − CT(ACTIN2). Last, relative levels in Col were set equal to 1 in all cases, and -fold change relative to Col was calculated. Primers are given in supplemental Table 1.
RESULTS
GTP Binding and Hydrolytic Activities of XLG2 Protein
The C-terminal regions of XLGs contain Gα domains (G-1 to G-5) (20, 21), which, in heterotrimeric G proteins, mediate GTP binding and hydrolysis (49). However, some amino acids conserved in the G-1 to G-5 domains of Gα proteins are not conserved within the corresponding domains of XLG proteins. Particularly, amino acids in the regions of the G-3 and G-5 domains, which are involved in Mg2+ coordination, GTP binding, and subsequent GTP hydrolysis, are not conserved (50) (Table 1). This raised the question of whether or not XLG proteins are able to bind and hydrolyze GTP. We addressed the biochemical properties of the three XLG proteins using GTP binding and GTPase assays. Results were similar for all three XLG proteins; results for XLG2 are reported here, and those for XLG1 and XLG3 are described in supplemental Figs. 1–4. To examine specific GTPγS binding and GTPase activities, we purified and assayed recombinant GPA1 and XLG proteins fused with His6 and TRX.
TABLE 1.
Conserved nucleotide binding motifs in selected G proteins
“At” indicates Arabidopsis thaliana, and “Os” indicates rice (Oryza sativa).
| Protein | G-1 | G-2 | G-3 | G-4 | G-5 |
|---|---|---|---|---|---|
| Consensus | GXXXXGK(S/T) | XXXTXXX | DXXGQ | NKXD | TCA |
| AtXLG1 | GNSGSGTS | EGVTSSS | RVPSR | NKYD | SKS |
| AtXLG2 | GSEKGGAT | EGLSSME | RLNPR | TKPD | VSL |
| AtXLG3 | GIEGSGTS | RVRTQGN | RVNAK | NKFD | ARA |
| AtGPA1 | GAGESGKS | RVRTTGV | DVGGQ | NKFD | TTA |
| OsRGA1 | GAGESGKS | RVRTNGV | DVGGQ | NKFD | TTA |
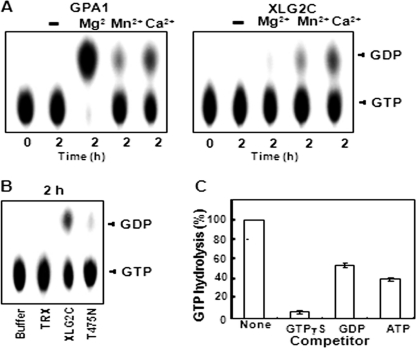
As expected, GPA1 bound GTPγS rapidly in the presence of Mg2+ (Fig. 1A), illustrating the known dependence of Gα proteins on this divalent cation (49). However, XLG2C did not bind GTPγS under these conditions (Fig. 1A). To address whether XLG2C has a preference for other divalent cations over Mg2+ for GTP binding activity, GTP binding filter assays were performed in the presence of Ca2+ instead of Mg2+. Interestingly, the presence of Ca2+ promoted XLG2C binding of GTPγS, whereas in the absence of any divalent ion, there was no GTPγS binding to XLG2C (Fig. 1B). By contrast, GPA1 showed weaker GTPγS binding in the presence of Ca2+ as compared with Mg2+ (Fig. 1C). Similar to XLG2C, XLG1 and XLG3 also bound GTPγS in the presence of Ca2+ (supplemental Figs. 3 and 4). These results indicate that XLG proteins prefer Ca2+ over Mg2+ as a cofactor for their GTP binding activities. To confirm the specificity of GTP binding, PCR-based mutagenesis was used to generate a mutation, T475N, in the G-1 domain of XLG2, a residue which is essential for binding to the phosphate moieties of guanine nucleotides (51). Similar mutations have been reported to abolish the GTP binding activity of other G proteins. For example, the T22N mutation of rice OsRab7 protein eliminates GTP binding (52, 53). Mutations S47C of mammalian Gαo and S48C of Gαi2 also impair GTPγS binding and create a dominant negative phenotype (54, 55). We designated this mutant protein XLG2C(T475N); this protein was also tagged with N-terminal His6 and TRX. GTP binding by XLG2C(T475N) protein was greatly reduced (Fig. 1C). As expected, purified His6 TRX control protein did not show any GTP binding activity (Fig. 1, A and C). Analogous mutations reduced the GTP binding of XLG1C and XLG3C (supplemental Figs. 3A and 4A).
FIGURE 1.
XLG2 has GTP binding activity in the presence of Ca2+. A, time course of [35S]GTPγS binding to GPA1 and lack of [35S]GTPγS binding to XLG2C in the presence of MgCl2. 100 nm XLG2C was incubated at 30 °C with 0.2 μm [35S]GTPγS in the presence of 10 mm MgCl2. 100 nm His6 TRX and 100 nm His6 TRX-tagged GPA1 were used as negative and positive controls, respectively. Aliquots (20 μl) were withdrawn at the indicated time points, filtered, and subjected to scintillation counting. B, time course of [35S]GTPγS binding to XLG2 in the presence of different concentrations of Ca2+. 100 nm XLG2C was incubated at 30 °C with 0.2 μm [35S]GTPγS in the presence or absence of 10 mm CaCl2. C, [35S]GTPγS binding to XLG2C, its mutant form XLG2C(T475N), and GPA1 in the presence of CaCl2 or MgCl2. The amount of [35S]GTPγS binding to XLG2C, XLG2C(T475N), and GPA1 was measured after a 2-h incubation in the presence of CaCl2 or MgCl2. Error bars, S.D.
Because heterotrimeric G protein α subunits, including Arabidopsis GPA1, have intrinsic GTPase activity, we examined the GTPase activity of XLG2C using thin layer chromatography (TLC) in the presence of various divalent cations. Although GPA1, like mammalian Gα subunits, exhibited the greatest hydrolytic activity in the presence of Mg2+, XLG2C showed little or no GTPase activity under this condition (Fig. 2A). In contrast, XLG2C GTPase activity was observed in the presence of Ca2+ (Fig. 2, A and B), although this activity was less than that of GPA1 in the presence of Mg2+ (Fig. 2A and supplemental Fig. 2C). As expected, no GTPase activity was observed for the mutant form of XLG2C, XLG2C(T475N) (Fig. 2B). The GTPase activity of XLG2C protein, which has no tag, was similar to that of His6 TRX-tagged XLG2C (supplemental Fig. 2B), indicating that His6 TRX tag does not affect the GTP binding and GTPase activities of XLG C terminus proteins.
FIGURE 2.
XLG2 has GTPase activity in the presence of Ca2+. A, GTPase activity of GPA1 and XLG2C in the presence of different divalent cofactors. GTPase activity was measured using the PEI-cellulose TLC plate method. B, GTPase activity of XLG2 and lack of GTPase activity of XLG2C(T475N) in the presence of CaCl2. The reactions were performed as described in A. Aliquots (2 μl) were withdrawn after incubation for 2 h and then separated by TLC. C, substrate competition test for XLG2 in the presence of CaCl2. Reactions were performed as described in A in the presence of competitors (3 μm unlabeled GTPγS, GDP, or ATP). The graph was drawn using quantitative TLC results (supplemental Fig. 2A) measured by ImageJ software. GTP hydrolysis was calculated as GDP/(GTP + GDP). GTP hydrolysis in the absence of competitor was designated as 100%. Error bars, S.D.
The specificity of the GTPase activity of XLG2C was also addressed (Fig. 2C). When a ∼100-fold excess of cold GTPγS, GDP, or ATP was added to the reaction, GTPγS had the greatest competitive effect, indicating that XLG2C specifically binds GTP (Fig. 2C). The full-length XLG2 protein exhibited only slightly increased GTPase activity compared with XLG2 C terminus protein (Table 2 and supplemental Fig. 2B), suggesting that the N-terminal region of XLG2 is dispensable for the GTPase activity. Similar results were observed for XLG1 and XLG3 (Table 2 and supplemental Figs. 3 and 4).
TABLE 2.
Comparison of GTPase activities of XLG proteins
GTPase activity calculated as the fraction of GTP remaining (GTP/(GTP + GDP)) after a 2-h incubation, as quantified by TLC. Smaller values indicate greater activity. Data shown are means ± S.D. of three independent replicates. ND, not determined. FL, full-length XLG proteins. C, C termini of XLG proteins, which include GTP binding domains.
| Mg2+ | Ca2+ | |
|---|---|---|
| XLG1 | ND | 0.18 ± 0.05 |
| XLG1C | 0.8992 ± 0.08 | 0.36 ± 0.06 |
| XLG2 | ND | 0.53 ± 0.11 |
| XLG2C | 0.9426 ± 0.03 | 0.68 ± 0.09 |
| XLG3 | ND | 0.33 ± 0.06 |
| XLG3C | 0.9482 ± 0.0 | 0.46 ± 0.08 |
RTV1 Is Identified as an XLG2-interacting Protein by Yeast Two-hybrid Screen
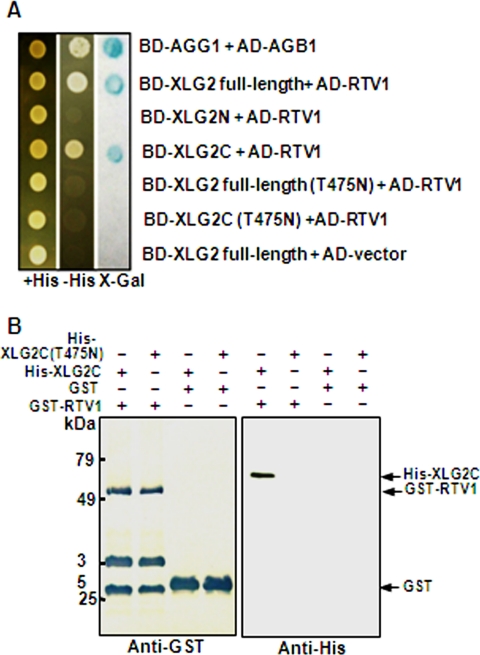
To address the functions of XLGs, we used a yeast two-hybrid screen to identify proteins that interact with XLG2. One XLG2-interacting protein recovered from the screen was RTV1 (related to vernalization 1) (At1g49480). RTV1 encodes a predicted protein of 226 amino acids with a plant-specific B3 DNA binding domain and a nuclear localization signal (NLS) (56). RTV1 shows an overall 81% identity with VRN1 in its amino acid sequence. VRN1 is a plant-specific B3-like DNA binding domain protein (57) that is required for maintenance of vernalization-mediated FLC repression and also promotes flowering in a vernalization-independent manner (58). To identify the domains of XLG2 that are responsible for RTV1 binding, we used partial cDNAs for additional yeast two-hybrid assays (Fig. 3A). Both full-length XLG2 and the C terminus of XLG2 interacted with RTV1, but the N terminus of XLG2 did not, indicating that the C terminus of XLG2 is responsible for the interaction in yeast.
FIGURE 3.
XLG2 but not XLG2(T475N) interacts with RTV1. A, yeast two-hybrid assay. β-Galactosidase activity was measured by the filter assay method (Invitrogen). The domains within XLG2 responsible for RTV1 binding were investigated. Wild type, domain deletion mutants, and a point mutant of XLG2 (T475N) were fused to the bait (BD) vector, and a full-length cDNA of RTV1 was fused to the prey (AD) vector. These constructs (described at the right) were co-transformed into pJG69-4A yeast cells, and their interactions were evaluated. AGG1 and AGB1 were used as a positive control for interaction (14). B, in vitro protein pull-down assay. Approximately 1 μg of GST or recombinant GST-tagged RTV1 was incubated with 1 μg of recombinant His-tagged AtXLG2C or AtXLG2C(T475N) and pulled down with GST resin after preblocking with 1% BSA. After five washes, the bound protein was fractionated by 12% SDS-PAGE and subjected to Western blot analysis with anti-GST or anti-His monoclonal antibodies. The extra bands may represent nonspecific cross-reactivity to bacterial proteins. Detection was by alkaline phosphatase/NBT directly on the blot membrane (left) or by enhanced chemiluminescence on x-ray film (right).
Interestingly, the XLG2 mutant proteins, XLG2 full-length (T475N) and XLG2C(T475N), which do not bind GTP, did not show any interaction with RTV1 in yeast (Fig. 3A), suggesting that XLG2 interacts with RTV1 in a GTP-dependent manner. These results were further confirmed by in vitro pull-down assays using recombinant proteins. We found that His6 TRX XLG2C but not His6 TRX XLG2C(T475N) binds to GST-RTV1 in vitro (Fig. 3B). Thus, the GTP binding activity of XLG2 appears to be required for its interaction with RTV1. Given the sequence similarity between RTV1 and VRN1, we also tested whether VRN1 interacts with XLGs using the yeast two-hybrid assay. However, VRN1 did not interact with any of the three XLGs (data not shown).
XLG2 and RTV1 Interact in Vivo and RTV1 Protein Localizes to Nucleus
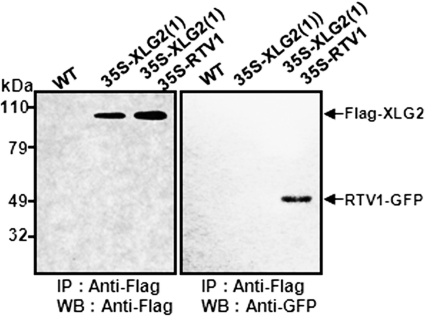
We used co-immunoprecipitation to test the protein-protein interaction between XLG2 and RTV1 in vivo (Fig. 4). Total proteins were extracted from wild type (non-transgenic), 35S-FLAG-XLG2(1), and 35S-FLAG-XLG2(1) 35S-RTV1-GFP double-overexpressing lines. Protein extracts were immunoprecipitated with anti-FLAG antibody as probe, and then protein gel blotting was performed to detect RTV1-GFP with anti-GFP antibody (Fig. 4). RTV1-GFP was detected in the proteins from 35S::FLAG-XLG2(1) 35S-RTV1-GFP double-overexpressing plants immunoprecipitated with anti-FLAG antibody. In control experiments, RTV1-GFP was not detected in the proteins immunoprecipitated with anti-FLAG antibody from WT or from 35S-FLAG-XLG2(1) plants, confirming the physical interaction of XLG2 with RTV1 in plant cells.
FIGURE 4.
XLG2 interacts with RTV1 in planta. Co-immunoprecipitation analysis is shown. Total proteins were extracted from WT and 35S-FLAG-XLG2(1) or 35S-FLAG-XLG2(1) 35S-RTV1-GFP overexpression lines and then immunoprecipitated with anti-FLAG antibody. The immunoprecipitates were separated by 10% SDS-PAGE, transferred to polyvinylidene difluoride membranes, and blotted with anti-FLAG (left) or anti-GFP (right) antibody. IP, immunoprecipitation; IB, immunoblot.
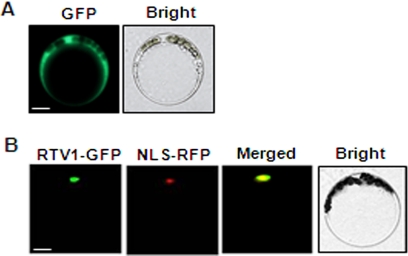
Given that XLG2 and RTV1 interact in planta, they would be expected to show identical or overlapping subcellular localization. All XLG proteins, including XLG2, have a nuclear localization sequence and localize to the nucleus, as observed in transient expression assays (21). We carried out transient expression assays to ascertain the localization of RTV1, which also has a predicted nuclear localization sequence (RKKK; amino acids 69–72). The green fluorescence signal from RTV1-GFP overlaps with the red fluorescence from the RFP-tagged NLS peptide, indicating that RTV1 localizes to the nucleus (Fig. 5).
FIGURE 5.
RTV1 localizes to the nucleus. A, Arabidopsis mesophyll protoplasts were transformed with GFP empty vector as a control. Bar, 10 μm. B, co-localization of RTV1-GFP with NLS-RFP (42). After co-transforming protoplasts with RTV1-GFP and NLS-RFP, the localization of these proteins was observed by fluorescence microscopy. Yellow regions in the merged image indicate overlap between the green and red fluorescent signals. Bar, 10 μm.
RTV1 Is Widely Expressed
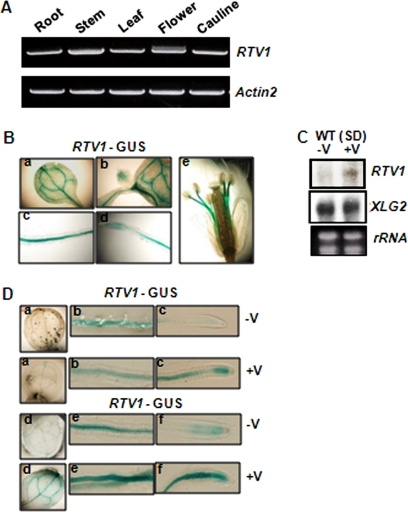
We applied several complementary approaches to assess RTV1 expression patterns. RT-PCR analysis showed that, similarly to XLG2 (21), RTV1 is expressed throughout the plant, including in roots, stems, leaves, flowers, and cauline leaves (Fig. 6A). We then used reporter gene analysis to examine expression patterns in more detail. Histochemical GUS staining of PRTV1::GUS lines showed that RTV1 is strongly expressed in cotyledon (a), shoot apical meristem (b), radicle (c), root tip (d), and staminar filaments (e) (Fig. 6B).
FIGURE 6.
RTV1 is expressed ubiquitously and its expression is enhanced by vernalization treatment. A, RTV1 expression in various organs as determined by RT-PCR. B, histochemical GUS staining of transgenic Arabidopsis expressing an RTV1 promoter-GUS fusion. a, cotyledon; b, shoot apical meristem; c and d, 7-day-old root; e, flower of 6-week-old plant. a and d, leaf; b and e, root; c and f, root tip. C, Northern blot analysis of RTV1 and XLG2 with (+V) or without (−V) 5 weeks of vernalization treatment. D, histochemical GUS staining of transgenic PRTV1-GUS line with (+V) or without (−V) 5 weeks of cold treatment. For GUS staining, a–c and d–f were incubated for 3 and 6 h, respectively.
Given the sequence similarity between VRN1 and RTV1 and thus a possible role of RTV1 in vernalization response, we also examined mRNA expression of RTV1 during the course of vernalization treatment (Fig. 6, C and D, and supplemental Fig. 5). RNA blot analysis showed that mRNA expression of RTV1, but not of XLG2, is increased after a 5-week cold treatment (Fig. 6C). Up-regulation of RTV1 by a 5-week cold treatment was also observed using PRTV1-GUS (Fig. 6D). Stronger expression of PRTV1-GUS was observed in plants that experienced a long term cold treatment (Fig. 6D, +V) compared with a control without cold treatment (Fig. 6D, −V).
To further dissect the increased level of RTV1 mRNA, we performed quantitative real-time RT-PCR using samples subjected to cold treatments ranging from 1 h to 5 weeks (supplemental Fig. 5). The mRNA level of RTV1 was rapidly increased almost 10-fold within 1 h of cold treatment and then decreased below the level of no cold treatment after 3 h of cold treatment. RTV1 mRNA expression eventually increased again ∼2-fold after a 5-week cold treatment (supplemental Fig. 5). XLG2 mRNA also exhibited the rapid increase within 1 h of cold treatment and the decrease after 3 h of cold treatment. Unlike RTV1, however, the level of XLG2 mRNA remained unchanged after a 5-week cold treatment (Fig. 6C and supplemental Fig. 5).
RTV1 Overexpression Promotes Early Flowering, Which Is Enhanced by Co-overexpression of XLG2
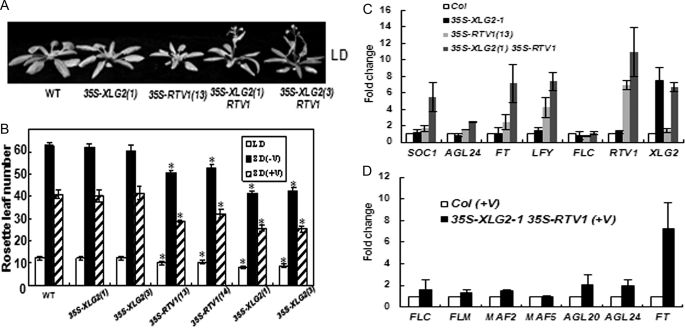
The ectopic expression of VRN1 causes early flowering due to the activation of FT and SOC1 (58). We accordingly examined flowering times of transgenic plants overexpressing RTV1. The 35S-FLAG-RTV1 plants exhibited rapid flowering under SD and a slight acceleration of flowering under LD (Fig. 7, A and B). In addition, when 35S-FLAG-RTV1 plants were vernalized, flowering was further accelerated (Fig. 7, A and B). Because XLG2 and RTV1 interact, we also evaluated the flowering time of 35S-FLAG-XLG2 35S-RTV1-GFP double overexpression lines. Interestingly, double overexpressor (Fig. 7, A and B, 35S-FLAG-XLG2 35S-RTV1-GFP) plants showed earlier flowering under LD and SD compared with both WT and 35S-FLAG-RTV1 plants, with or without vernalization treatment (Fig. 7B). Thus, the interaction between XLG2 and RTV1 appears to enhance RTV1 activity in vivo. To investigate the molecular basis of this early flowering phenotype, we examined the expression of floral pathway genes in 35S-FLAG-XLG2 and 35S-FLAG-RTV1 overexpression lines and in 35S-FLAG-XLG2 35S-RTV1-GFP double overexpression lines under SD using quantitative RT-PCR. Introduction of 35S-FLAG-XLG2 alone did not promote transcript levels of the floral pathway genes SOC1, AGL24, FT, and LFY (Fig. 7C). However, the abundance of these transcripts was increased in 35S-FLAG-RTV1 plants and even more so in 35S-FLAG-XLG2 35S-RTV1-GFP plants (Fig. 7C).
FIGURE 7.
35S-FLAG-RTV1 plants flower early in both LD and SD, and earlier flowering is further enhanced in 35S-FLAG-XLG2 35S-RTV1-GFP plants. A, early flowering of 35S-FLAG-XLG2 35S-RTV1-GFP in LD. B, flowering times were determined as total rosette leaf number at the time of flowering under LD and SD conditions with (+V) or without (−V) 5 weeks of cold treatment. At least 12 plants of each genotype were used. Statistical significance was determined by Student's t test (*, p < 0.001 versus Col). C, all transcript levels were normalized to ACTIN2 levels using the formula, CT = CT(each gene) − CT(ACTIN2). -Fold changes are relative to results from wild type (Col). Real-time RT-PCR was performed with cDNAs from 2-week-old wild type and transgenic plants grown in SD conditions. Each bar represents an average of three independent replicate experiments. D, the expression of FLC, FLC clade (MAFs), and floral integrator genes in 35S-FLAG-XLG2 35S-RTV1-GFP plants after 5 weeks of cold treatment (+V). All results were obtained as described in C.
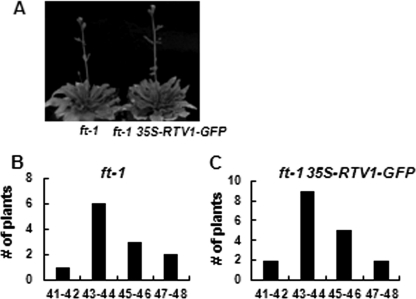
Vernalization triggers the repression of FLC and other FLC clade genes (37, 59). mRNA levels of FLC and its clade genes MAF1, MAF2, and MAF5 in 35S-FLAG-RTV1 and 35S-FLAG-XLG2 35S-RTV1-GFP plants immediately after vernalization showed no significant difference compared with vernalized wild type in the extent of this repression (i.e. -fold change compared with Col was <2). After vernalization, however, the floral pathway integrator genes SOC1 and AGL24 and particularly the levels of FT mRNA were higher in these lines than in Col (i.e. -fold change compared with Col was >2) (Fig. 7D). To analyze the effect of ft mutation (60) on the flowering time of 35S-RTV1-GFP plants, 35S-RTV1-GFP construct was transformed into the ft mutant, and flowering times were determined (Fig. 8). 35S-RTV1-GFP in the ft mutant background had the same late flowering phenotype as ft mutants (Fig. 8), indicating that early flowering of 35S::RTV1 requires the elevated expression of FT. Therefore, accelerated flowering of overexpressing XLG2 and RTV1 plants (with or without vernalization) apparently occurs directly through regulation of the floral pathway integrator gene, FT, through a mechanism independent of repression of FLC and its clade genes.
FIGURE 8.
Flowering phenotypes of 35S-RTV1 ft-1. A, 35S-RTV1 ft-1 flowering is similar to ft-1. B and C, data represent plant frequency distributions based on rosette leaf number at flowering under LD ft-1 (n = 12) (B) and 35S-RTV1 ft-1 (n = 18 T1 individuals) (C). Error bars, S.D.
RTV1 Binds DNA and XLG2 Enhances This DNA Binding Activity
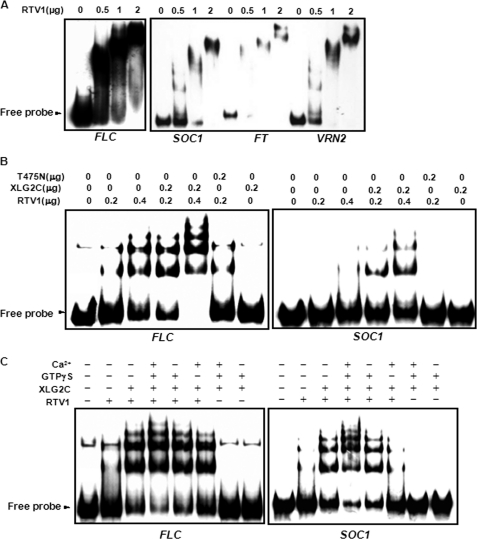
VRN1 shows a non-sequence-specific DNA binding activity in vitro (58). Given the sequence similarity between VRN1 and RTV1 and the nuclear localization of RTV1, we examined the DNA binding activity of recombinant RTV1 protein. EMSAs were performed using DNA fragments derived from promoters of FLC, SOC1, FT, VRN2, or DFR (dihydroflavonol 4-reductase). Recombinant RTV1 protein bound to all DNA fragments tested (Fig. 9A and supplemental Fig. 6). These DNA fragments do not share any common sequence motif, illustrating that, similar to VRN1, RTV1 has non-sequence-specific DNA binding activity, at least in these in vitro assays. The interaction between XLG2 and RTV1 prompted us to hypothesize that this interaction may modulate the DNA binding activity of RTV1. Accordingly, recombinant XLG2 C-terminal domain protein (XLG2C), which contains the RTV1 interaction domains (Fig. 3), was added in the reaction with RTV1, and the binding strength was evaluated by the amount of probe retardation (Fig. 9B). The addition of XLG2C increased the DNA binding activity of RTV1 (Fig. 9B), whereas the mutant protein XLG2C(T475N), which fails to interact with RTV1, did not (Fig. 8B). Because XLG2C(T475N) mutant protein also fails to bind GTP in vitro (Fig. 1C), we examined the effect of GTP binding of XLG2C on the DNA binding activity of RTV1. XLG2C, GTPγS, and Ca2+ were added in the reaction together with RTV1. Under these conditions, XLG2C formed a supercomplex with RTV1 and probes, suggesting that activation of XLG2C by GTP binding contributes to the enhanced DNA binding of RTV1 in vitro (Fig. 9C).
FIGURE 9.
RTV1 has non-sequence-specific DNA binding activity in vitro that is enhanced by XLG2. A, EMSA of RTV1 using FLC, SOC1, FT, or VRN2 promoter regions. RTV1 protein (0–57 nm; 0–2 μg) and 400-bp radiolabeled double-stranded FLC, SOC1, FT, or VRN2 probe were incubated and then separated on a 5% Tris-borate EDTA-polyacrylamide gel. B, increased DNA binding by RTV1 in the presence of XLG2C but not in the presence of XLG2C(T475N). The reaction was performed as described in A together with 50 nm XLG2C or XLG2C(T475N) using FLC or SOC1 probes, respectively. C, the effect of GTPγS binding by XLG2 in the presence of Ca2+. The reaction was performed as described in B together with 1 mm GTPγS and 10 mm Ca2+ using FLC or SOC1 probes, respectively.
RTV1 Is Associated with FT and SOC1 Promoters in Vivo
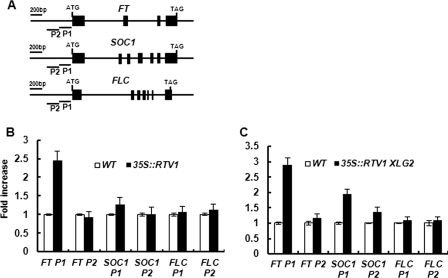
We showed that RTV1 overexpression lines show early flowering due to increased expression of floral pathway integrator genes independent of regulation of FLC expression (Figs. 7 and 8) and that RTV1 binds to DNA in a non-sequence-specific manner in vitro (Fig. 9A and supplemental Fig. 9). To address whether RTV1 binds to promoters of floral pathway integrator genes, such as FT and SOC1, in vivo, ChIP assays were employed. For these assays, wild type, 35S-FLAG-RTV1(13), and 35S-FLAG-XLG2(1) 35S-RTV1-GFP double-overexpresssing lines were used. After immunoprecipitation with anti-FLAG or anti-GFP antibodies, enrichment of FT and SOC1 promoter regions was assayed by quantitative PCR. As shown in Fig. 10, B and C, enrichment of the FT promoter region, 1–200 bp upstream from the start codon (P1) was observed in the 35S-FLAG-RTV1(13) line, with slightly enhanced enrichment in the 35S-FLAG-XLG2(1) 35S-RTV1-GFP overexpression lines. In addition, SOC1 promoter regions were not enriched in 35S-FLAG-RTV1(13), whereas enrichment of the SOC1 promoter region, 1–200 bp upstream from the start codon (P1) was observed from the 35S-FLAG-XLG2(1) 35S-RTV1-GFP (Fig. 10C). No enrichment of FLC regions was observed in either the 35S-FLAG-RTV1(13) line or the 35S-FLAG-XLG2(1) 35S-RTV1-GFP line (Fig. 10, B and C). Taken together, our results indicate that XLG2 promotes the DNA binding activity of RTV1 specifically to promoter regions of FT and SOC1 in vivo and thus leads to the activation of floral integrator genes (Fig. 11).
FIGURE 10.
RTV1 is associated with FT and SOC1 chromatin at their promoter regions. A, schematic diagram of the FT, SOC1, and FLC genomic regions. Exons are represented by black boxes, whereas introns and upstream regions are represented by black lines. Primer regions used in ChIP assays are designated as P1 and P2. B and C, ChIP-quantitative PCR analyses. WT, 35S-FLAG-RTV1(13) (B), and 35S-FLAG-XLG2(1) 35S-RTV1-GFP (C) transgenic seedlings were grown under SD. Chromatin was isolated with extraction buffers. Anti-FLAG or anti-GFP antibodies were used for immunoprecipitation. Precipitated chromatin from ChIP assays from WT and transgenic seedlings was quantified by quantitative PCR with specific primers for regions indicated in A. Error bars, S.D.
FIGURE 11.
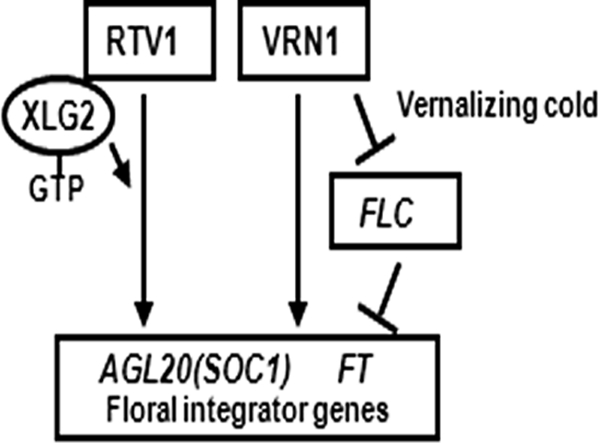
Model for the roles of RTV1 and XLG2 in the regulation of floral integrator genes in Arabidopsis. The GTP-bound form of XLG2 promotes RTV1 DNA binding to floral integrator genes, FT and SOC1, leading to early flowering.
DISCUSSION
XLG Proteins Are Specific GTPases
Here we report new biochemical and functional properties of the XLG proteins. Conventional Gα proteins have highly conserved domains involved in GTP binding and hydrolysis. Because of variations in amino acid sequences within these domains in the three XLG proteins (Table 1), it was not clear whether or not the XLG proteins function as GTPases. XLG1 had been shown previously to bind GTP (20), but GTP binding of XLG2 and XLG3, or GTPase activity of any of the XLG proteins had not been evaluated. In this study, we show that the three XLG proteins indeed specifically bind GTP and possess GTPase activity. Thus, we provide biochemical evidence that the XLG proteins, despite their unusual structure, are bona fide G proteins. Given the variations in amino acid sequences of the XLG GTP-binding domains, we initially hypothesized that the XLG-specific N termini might provide structural scaffolds that enable GTP binding. However, this appears not to be the case because the C termini of the XLGs alone are sufficient for GTP binding and GTPase activity (Fig. 1 and supplemental Figs. 2–4). The specific functions of the unique N termini of the XLG proteins remain to be uncovered. XLG2, which is the most divergent of the three XLGs in the G-1, G-2, G-3, and G-4 domains, also has the slowest GTPase activity (Table 2). Other types of G proteins, such as Ras and EF-Tu (61), associate with GTPase-activating proteins (GAPs) (62). It will be interesting in the future to determine whether GAPs exist for the XLGs.
The XLGs are distinct from canonical Gα subunits in their dependence on Ca2+, rather than Mg2+, as a cofactor. The G-1 to G-3 domains of G proteins provide critical contacts for the α-, β-, and γ-phosphates of the guanine nucleotide and also for the coordination of Mg2+ (50). The G-4 and G-5 domains are important for the binding of the guanine ring (50). XLG2 has the conserved amino acid motifs in the G-1, G-2, and G-4 regions. XLG2 utilizes Ca2+ but not Mg2+ as a cofactor although it has the well conserved threonine residue in the G-2 region (Table 1), which plays a critical role in coordinating Mg2+ (50). However, XLG proteins do not have the conserved motifs of the G-3 and G-5 regions (21) (Table 1). The glutamine residue in the G-3 (DXXGQ) domain is involved in coordination of Mg2+ mediated by water, and the main chain of the glycine residue forms hydrogen bonds with the γ-phosphate of GTP (49, 51). When introduced in Ras, virtually all mutations in the G-3 domain reduce its GTPase activity and disrupt Ras binding to Ras-GAP (63). Corresponding mutations in Gαs (64, 65) and in Gαi1 (66) also reduce the GTPase activity. In addition, mutation at the G-5 (TCA) alanine in Gαi promotes rapid GDP dissociation, although the intrinsic GTPase activity of Gαi is not changed (67). Taken together, variations in amino acid sequences of XLG2 and the other two XLG proteins probably contribute to their different cofactor requirement.
In summary, our data demonstrate that all three XLG proteins exhibit both GTP binding and GTPase activity. Our results also revealed an interesting divergence from Gα proteins because XLG proteins cannot utilize Mg2+ as a cofactor. The unusual requirement for Ca2+ in XLG protein GTP binding and hydrolysis raises the possibility that XLG proteins may be involved in the regulation of a variety of physiological and metabolic processes that are dependent on internal Ca2+ concentration, a well known component in many signal transduction chains in plants and other eukaryotes (68).
RTV1 Is an XLG2-interacting Protein
RTV1 was identified as an XLG2-interacting protein by a yeast two-hybrid screen using XLG2 as bait (Fig. 3A). We confirmed the interaction between RTV1 and XLG2 in vitro and in planta (Fig. 3B and Fig. 4), indicating that RTV1 is a bona fide interacting protein of XLG2. Interestingly, the Gα-like C terminus of XLG2, rather than the N terminus, interacts with RTV1 (Fig. 3A). The interaction is not observed with the XLG2C(T475N) mutant (Fig. 3, A and B), which lacks GTP binding activity (Fig. 1C). This suggests that GTP binding by XLG2 plays an important role in protein-protein interaction.
The RTV1 gene encodes a protein with a B3-like DNA binding domain, a domain that is unique to plants (56). The expression of RTV1 in vasculature is similar to that of other floral regulators, such as FT (69, 70), establishing an overlapping expression profile between RTV1 and the RTV1 target, FT, and suggesting that RTV1 is capable of regulating FT in locations where FT is expressed. The expression pattern of RTV1 and the nuclear localization of RTV1 are also very similar to those of XLG2 (Fig. 5 and 6) (21), consistent with in planta interaction of the proteins.
RTV1 Promotes Floral Integrator Gene Expression and Flowering via an XLG2-related Mechanism
Overexpression of RTV1 led to early flowering (Fig. 7) through increased expression of floral integrator genes, such as FT and SOC1 (Fig. 7C). Consistent with this model, introgression of 35S::RTV1 into an ft background eliminated the early flowering phenotype (Fig. 8). Moreover, when RTV1 was overexpressed together with XLG2, the expression of the floral integrator genes was further enhanced, and these plants showed even earlier flowering compared with RTV1-only-overexpressing lines (Fig. 7). These results lead to a model whereby XLG2 interaction with RTV1 promotes RTV1 activity, leading to enhanced expression of the floral pathway integrator genes (Fig. 11).
XLG2 Promotes RTV1 Binding to Promoter Regions of Floral Integrator Genes
RTV1 exhibits strong DNA binding activities in vitro with DNA fragments derived from promoter regions of FLC, SOC1, FT, and VRN2 (Fig. 9) as well as with the promoter of the non-flowering related gene DRF (supplemental Fig. 6). Such non-sequence-specific binding to DNA fragments in vitro was also observed for VRN1, which also belongs to the B3 domain superfamily (58).
Swaminathan et al. (56) classified B3 genes into four families that are typified by ABI3 (abscisic acid-insensitive 3 (71)), ARF1 (auxin-responsive factor 1 (72)), RAV1 (related to ABI3/VP1 (73)), and REM (reproductive meristem). The 76 REM genes in Arabidopsis have been divided into six subgroups (A–F) based on phylogenetic analysis (56). RTV1 and VRN1 belong to the six genes of subgroup A of the REM family. VRN1 has been the only REM gene with known function (58). Unlike other B3 domain proteins, which show sequence-specific DNA binding (e.g. ABI3, RAV1, and auxin-response factors), both VRN1 and RTV1 bind to DNA in a non-sequence-specific manner in vitro (58). It is interesting to note that XLG2C enhances the DNA binding activity of RTV1 (Fig. 9, B and C). XLG2 in the presence of Ca2+ and GTPγS enhances the DNA binding activity of RTV1 even further (Fig. 9C), whereas the T475N mutant of XLG2C, which does not bind GTP, has no effect on the DNA binding activity of RTV1 (Fig. 9B). Taken together, these results suggest that XLG2 activation caused by GTP binding in the presence of Ca2+ enhances the DNA binding activity of RTV1.
Although we did not observe sequence-specific DNA binding of RTV1 in vitro, specificity may be conferred by other components, such as other interacting proteins, present in planta but not in vitro. In fact, our ChIP analyses showed that RTV1 binds only to certain target genes in planta. In ChIP assays using 10-day-old 35S-FLAG-RTV1(13) and 35S-FLAG-XLG2(1) 35S-RTV1-GFP seedlings, we consistently found that RTV1 directly interacts with FT and SOC1 promoters. Moreover, a slight increase of RTV1 at the FT promoter and an enrichment of RTV1 at the SOC1 promoter were observed in 35S-FLAG-XLG2(1) 35S-RTV1-GFP seedlings compared with 35S-FLAG-RTV1(13) (Fig. 10, B and C). This suggests that XLG2 facilitates RTV1 activation, which then results in transcriptional activation of FT and SOC1, an interpretation that is also consistent with our in vitro gel shift assays (Fig. 9C) and our gene expression assays (Fig. 7).
We did not observe any significant enrichment of RTV1 at FLC regions by ChIP assays (Fig. 10, B and C); nor did we observe any enhanced repression of FLC upon vernalization of 35S-FLAG-XLG2(1) 35S-RTV1-GFP lines (Fig. 7D). This suggests that RTV1 and XLG2 overexpression directly activate floral integrator genes in an FLC-independent manner. VRN1, which shares significant sequence homology with RTV1, also activates floral integrator genes via an FLC-independent pathway (58), and ectopic expression of RTV1 also accelerates flowering. These two proteins differ, however, in that VRN1, but not RTV1, has an additional function in stabilizing FLC repression postvernalization (58), and RTV1, but not VRN1, binds and is regulated by XLG2 (Fig. 11).
In summary, our work provides new insight into signaling by Ca2+ and G proteins. We show that RTV1 and XLG2 function together in the regulation of flowering time. FT and SOC1 are direct targets of RTV1. It is likely that GTP-bound XLG2 and/or XLG2 cycling between GTP- and GDP-bound states, which occur in the presence of Ca2+, are required for enhanced RTV1 association with targets such as FT and SOC1. Circadian oscillations in intracellular free calcium ions ([Ca2+]i) occur and encode photoperiodic information (74, 75). It is interesting to note that XLG2 and RTV1 transcripts exhibit in-phase circadian oscillation (see the Diurnal Search Tool Web site). It is tantalizing to speculate that XLG2-RTV1 mediates some of the outputs from the oscillations in [Ca2+]i. However, the role of [Ca2+]i oscillation in flowering time control is poorly understood. Further studies on the regulatory network of Ca2+, XLG2, and RTV1 should provide interesting insights into the roles of XLG proteins in Ca2+-mediated signaling pathways.
Supplementary Material
This research was supported by United States Department of Agriculture Grant 2003-35304-13924 and Penn State funding (to S. M. A.), by National Science Foundation Grant IOS 0950785 (to S. S.), and Next-Generation BioGreen 21 Program SSAC Grant PJ008087 and a National Research Foundation of Korea Grant funded by the Korean Government (MEST Grant 2011-0013137) (to J. B. H.).

This article contains supplemental Table 1 and Figs. 1–6.
- LD
- long day
- SD
- short day
- GUS
- β-glucuronidase
- TRX
- thioredoxin
- GAP
- GTPase-activating protein
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate
- SC
- synthetic complete
- NLS
- nuclear localization signal.
REFERENCES
- 1. Hepler J. R., Gilman A. G. (1992) G proteins. Trends Biochem. Sci. 17, 383–387 [DOI] [PubMed] [Google Scholar]
- 2. Assmann S. M. (2002) Heterotrimeric and unconventional GTP binding proteins in plant cell signaling. Plant Cell 14, S355–S373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jones A. M., Assmann S. M. (2004) Plants: the latest model system for 6-protein research. EMBO Rep. 5, S572–S578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ma H. (1994) GTP-binding proteins in plants. New members of an old family. Plant Mol. Biol. 26, 1611–1636 [DOI] [PubMed] [Google Scholar]
- 5. Gilman A. G. (1987) G proteins. Transducers of receptor-generated signals. Annu. Rev. Biochem. 56, 615–649 [DOI] [PubMed] [Google Scholar]
- 6. Gutkind J. S. (1998) The pathways connecting G protein-coupled receptors to the nucleus through divergent mitogen-activated protein kinase cascades. J. Biol. Chem. 273, 1839–1842 [DOI] [PubMed] [Google Scholar]
- 7. Hamm H. E. (1998) The many faces of G protein signaling. J. Biol. Chem. 273, 669–672 [DOI] [PubMed] [Google Scholar]
- 8. McCudden C. R., Hains M. D., Kimple R. J., Siderovski D. P., Willard F. S. (2005) G-protein signaling. Back to the future. Cell Mol. Life Sci. 62, 551–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Assmann S. M. (2004) Plant G proteins, phytohormones, and plasticity. Three questions and a speculation. Sci. STKE 2004, re20. [DOI] [PubMed] [Google Scholar]
- 10. Temple B. R., Jones A. M. (2007) The plant heterotrimeric G-protein complex. Annu. Rev. Plant Biol. 58, 249–266 [DOI] [PubMed] [Google Scholar]
- 11. Assmann S. M. (2005) G proteins Go green. A plant G protein signaling FAQ sheet. Science 310, 71–73 [DOI] [PubMed] [Google Scholar]
- 12. Ma H., Yanofsky M. F., Meyerowitz E. M. (1990) Molecular cloning and characterization of GPA1, a G protein α subunit gene from Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 87, 3821–3825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Weiss C. A., Garnaat C. W., Mukai K., Hu Y., Ma H. (1994) Isolation of cDNAs encoding guanine nucleotide-binding protein β-subunit homologues from maize (ZGB1) and Arabidopsis (AGB1). Proc. Natl. Acad. Sci. U.S.A. 91, 9554–9558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mason M. G., Botella J. R. (2000) Completing the heterotrimer. Isolation and characterization of an Arabidopsis thaliana G protein γ-subunit cDNA. Proc. Natl. Acad. Sci. U.S.A. 97, 14784–14788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mason M. G., Botella J. R. (2001) Isolation of a novel G-protein γ-subunit from Arabidopsis thaliana and its interaction with Gβ. Biochim. Biophys. Acta 1520, 147–153 [DOI] [PubMed] [Google Scholar]
- 16. Gookin T. E., Kim J., Assmann S. M. (2008) Whole proteome identification of plant candidate G-protein coupled receptors in Arabidopsis, rice, and poplar. Computational prediction and in vivo protein coupling. Genome Biol. 9, R120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moriyama E. N., Strope P. K., Opiyo S. O., Chen Z., Jones A. M. (2006) Mining the Arabidopsis thaliana genome for highly divergent seven-transmembrane receptors. Genome Biol. 7, R96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chakravorty D., Trusov Y., Zhang W., Acharya B. R., Sheahan M. B., McCurdy D. W., Assmann S. M., Botella J. R. (2011) An atypical heterotrimeric G-protein γ-subunit is involved in guard cell K+-channel regulation and morphological development in Arabidopsis thaliana. Plant J. 67, 840–851 [DOI] [PubMed] [Google Scholar]
- 19. Perfus-Barbeoch L., Jones A. M., Assmann S. M. (2004) Plant heterotrimeric G protein function. Insights from Arabidopsis and rice mutants. Curr. Opin. Plant Biol. 7, 719–731 [DOI] [PubMed] [Google Scholar]
- 20. Lee Y. R., Assmann S. M. (1999) Arabidopsis thaliana “extra-large GTP-binding protein” (AtXLG1). A new class of G-protein. Plant Mol. Biol. 40, 55–64 [DOI] [PubMed] [Google Scholar]
- 21. Ding L., Pandey S., Assmann S. M. (2008) Arabidopsis extra-large G proteins (XLGs) regulate root morphogenesis. Plant J. 53, 248–263 [DOI] [PubMed] [Google Scholar]
- 22. Pandey S., Monshausen G. B., Ding L., Assmann S. M. (2008) Regulation of root-wave response by extra large and conventional G proteins in Arabidopsis thaliana. Plant J. 55, 311–322 [DOI] [PubMed] [Google Scholar]
- 23. Zhu H., Li G. J., Ding L., Cui X., Berg H., Assmann S. M., Xia Y. (2009) Arabidopsis extra large G-protein 2 (XLG2) interacts with the Gβ subunit of heterotrimeric G protein and functions in disease resistance. Mol. Plant 2, 513–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Parcy F. (2005) Flowering. A time for integration. Int. J. Dev. Biol. 49, 585–593 [DOI] [PubMed] [Google Scholar]
- 25. Moon J., Lee H., Kim M., Lee I. (2005) Analysis of flowering pathway integrators in Arabidopsis. Plant Cell Physiol. 46, 292–299 [DOI] [PubMed] [Google Scholar]
- 26. Michael T. P., Salomé P. A., Yu H. J., Spencer T. R., Sharp E. L., McPeek M. A., Alonso J. M., Ecker J. R., McClung C. R. (2003) Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science 302, 1049–1053 [DOI] [PubMed] [Google Scholar]
- 27. Liu C., Chen H., Er H. L., Soo H. M., Kumar P. P., Han J. H., Liou Y. C., Yu H. (2008) Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 135, 1481–1491 [DOI] [PubMed] [Google Scholar]
- 28. Zeevaart J. A. (2008) Leaf-produced floral signals. Curr. Opin. Plant Biol. 11, 541–547 [DOI] [PubMed] [Google Scholar]
- 29. Turck F., Fornara F., Coupland G. (2008) Regulation and identity of florigen. FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 59, 573–594 [DOI] [PubMed] [Google Scholar]
- 30. Abe M., Kobayashi Y., Yamamoto S., Daimon Y., Yamaguchi A., Ikeda Y., Ichinoki H., Notaguchi M., Goto K., Araki T. (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056 [DOI] [PubMed] [Google Scholar]
- 31. Melzer S., Lens F., Gennen J., Vanneste S., Rohde A., Beeckman T. (2008) Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana. Nat. Genet. 40, 1489–1492 [DOI] [PubMed] [Google Scholar]
- 32. Samach A., Onouchi H., Gold S. E., Ditta G. S., Schwarz-Sommer Z., Yanofsky M. F., Coupland G. (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288, 1613–1616 [DOI] [PubMed] [Google Scholar]
- 33. Hepworth S. R., Valverde F., Ravenscroft D., Mouradov A., Coupland G. (2002) Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J. 21, 4327–4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sung S., Amasino R. M. (2004) Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427, 159–164 [DOI] [PubMed] [Google Scholar]
- 35. Michaels S. D., Amasino R. M. (2001) Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13, 935–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bastow R., Mylne J. S., Lister C., Lippman Z., Martienssen R. A., Dean C. (2004) Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427, 164–167 [DOI] [PubMed] [Google Scholar]
- 37. Sung S., He Y., Eshoo T. W., Tamada Y., Johnson L., Nakahigashi K., Goto K., Jacobsen S. E., Amasino R. M. (2006) Epigenetic maintenance of the vernalized state in Arabidopsis thaliana requires LIKE HETEROCHROMATIN PROTEIN 1. Nat. Genet. 38, 706–710 [DOI] [PubMed] [Google Scholar]
- 38. Northup J. K., Smigel M. D., Gilman A. G. (1982) The guanine nucleotide activating site of the regulatory component of adenylate cyclase. Identification by ligand binding. J. Biol. Chem. 257, 11416–11423 [PubMed] [Google Scholar]
- 39. Smigel M. D., Northup J. K., Gilman A. G. (1982) Characteristics of the guanine nucleotide-binding regulatory component of adenylate cyclase. Recent Prog. Horm. Res. 38, 601–624 [DOI] [PubMed] [Google Scholar]
- 40. Heo J. B., Rho H. S., Kim S. W., Hwang S. M., Kwon H. J., Nahm M. Y., Bang W. Y., Bahk J. D. (2005) OsGAP1 functions as a positive regulator of OsRab11-mediated TGN to PM or vacuole trafficking. Plant Cell Physiol. 46, 2005–2018 [DOI] [PubMed] [Google Scholar]
- 41. Benfey P. N., Chua N. H. (1990) The cauliflower mosaic virus 35S promoter. Combinatorial regulation of transcription in plants. Science 250, 959–966 [DOI] [PubMed] [Google Scholar]
- 42. Cheong Y. H., Moon B. C., Kim J. K., Kim C. Y., Kim M. C., Kim I. H., Park C. Y., Kim J. C., Park B. O., Koo S. C., Yoon H. W., Chung W. S., Lim C. O., Lee S. Y., Cho M. J. (2003) BWMK1, a rice mitogen-activated protein kinase, locates in the nucleus and mediates pathogenesis-related gene expression by activation of a transcription factor. Plant Physiol. 132, 1961–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Karimi M., De Meyer B., Hilson P. (2005) Modular cloning in plant cells. Trends Plant Sci. 10, 103–105 [DOI] [PubMed] [Google Scholar]
- 44. Clough S. J., Bent A. F. (1998) Floral dip. A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 [DOI] [PubMed] [Google Scholar]
- 45. Despres C., Subramaniam R., Matton D. P., Brisson N. (1995) The activation of the potato PR-10a gene requires the phosphorylation of the nuclear factor PBF-1. Plant Cell 7, 589–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Johnson K. D., Bresnick E. H. (2002) Dissecting long-range transcriptional mechanisms by chromatin immunoprecipitation. Methods 26, 27–36 [DOI] [PubMed] [Google Scholar]
- 47. Heo J. B., Sung S. (2011) Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331, 76–79 [DOI] [PubMed] [Google Scholar]
- 48. Gendrel A. V., Lippman Z., Martienssen R., Colot V. (2005) Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods 2, 213–218 [DOI] [PubMed] [Google Scholar]
- 49. Sprang S. R. (1997) G protein mechanisms. Insights from structural analysis. Annu. Rev. Biochem. 66, 639–678 [DOI] [PubMed] [Google Scholar]
- 50. Barren B., Artemyev N. O. (2007) Mechanisms of dominant negative G-protein α subunits. J. Neurosci. Res. 85, 3505–3514 [DOI] [PubMed] [Google Scholar]
- 51. Noel J. P., Hamm H. E., Sigler P. B. (1993) The 2.2 Å crystal structure of transducin-α complexed with GTPγS. Nature 366, 654–663 [DOI] [PubMed] [Google Scholar]
- 52. Nahm M. Y., Kim S. W., Yun D., Lee S. Y., Cho M. J., Bahk J. D. (2003) Molecular and biochemical analyses of OsRab7, a rice Rab7 homolog. Plant Cell Physiol. 44, 1341–1349 [DOI] [PubMed] [Google Scholar]
- 53. Wong K. A., Lodish H. F. (2006) A revised model for AMP-activated protein kinase structure. The α-subunit binds to both the β- and γ-subunits although there is no direct binding between the β- and γ-subunits. J. Biol. Chem. 281, 36434–36442 [DOI] [PubMed] [Google Scholar]
- 54. Slepak V. Z., Katz A., Simon M. I. (1995) Functional analysis of a dominant negative mutant of Gαi2. J. Biol. Chem. 270, 4037–4041 [DOI] [PubMed] [Google Scholar]
- 55. Slepak V. Z., Wilkie T. M., Simon M. I. (1993) Mutational analysis of G protein α subunit Go α expressed in Escherichia coli. J. Biol. Chem. 268, 1414–1423 [PubMed] [Google Scholar]
- 56. Swaminathan K., Peterson K., Jack T. (2008) The plant B3 superfamily. Trends Plant Sci. 13, 647–655 [DOI] [PubMed] [Google Scholar]
- 57. Waltner J. K., Peterson F. C., Lytle B. L., Volkman B. F. (2005) Structure of the B3 domain from Arabidopsis thaliana protein At1g16640. Protein Sci. 14, 2478–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Levy Y. Y., Mesnage S., Mylne J. S., Gendall A. R., Dean C. (2002) Multiple roles of Arabidopsis VRN1 in vernalization and flowering time control. Science 297, 243–246 [DOI] [PubMed] [Google Scholar]
- 59. Ratcliffe O. J., Kumimoto R. W., Wong B. J., Riechmann J. L. (2003) Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family. MAF2 prevents vernalization by short periods of cold. Plant Cell 15, 1159–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chiang G. C., Barua D., Kramer E. M., Amasino R. M., Donohue K. (2009) Major flowering time gene, flowering locus C, regulates seed germination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 106, 11661–11666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Eccleston J. F., Moore K. J., Morgan L., Skinner R. H., Lowe P. N. (1993) Kinetics of interaction between normal and proline 12 Ras and the GTPase-activating proteins, p120-GAP and neurofibromin. The significance of the intrinsic GTPase rate in determining the transforming ability of ras. J. Biol. Chem. 268, 27012–27019 [PubMed] [Google Scholar]
- 62. Scheffzek K., Ahmadian M. R., Wiesmüller L., Kabsch W., Stege P., Schmitz F., Wittinghofer A. (1998) Structural analysis of the GAP-related domain from neurofibromin and its implications. EMBO J. 17, 4313–4327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Frech M., Darden T. A., Pedersen L. G., Foley C. K., Charifson P. S., Anderson M. W., Wittinghofer A. (1994) Role of glutamine 61 in the hydrolysis of GTP by p21H-ras. An experimental and theoretical study. Biochemistry 33, 3237–3244 [DOI] [PubMed] [Google Scholar]
- 64. Landis C. A., Masters S. B., Spada A., Pace A. M., Bourne H. R., Vallar L. (1989) GTPase-inhibiting mutations activate the α chain of Gs and stimulate adenylyl cyclase in human pituitary tumors. Nature 340, 692–696 [DOI] [PubMed] [Google Scholar]
- 65. Graziano M. P., Gilman A. G. (1989) Synthesis in Escherichia coli of GTPase-deficient mutants of Gs α. J. Biol. Chem. 264, 15475–15482 [PubMed] [Google Scholar]
- 66. Coleman D. E., Berghuis A. M., Lee E., Linder M. E., Gilman A. G., Sprang S. R. (1994) Structures of active conformations of Gi α 1 and the mechanism of GTP hydrolysis. Science 265, 1405–1412 [DOI] [PubMed] [Google Scholar]
- 67. Posner B. A., Mixon M. B., Wall M. A., Sprang S. R., Gilman A. G. (1998) The A326S mutant of Giα1 as an approximation of the receptor-bound state. J. Biol. Chem. 273, 21752–21758 [DOI] [PubMed] [Google Scholar]
- 68. Reddy V. S., Reddy A. S. (2004) Proteomics of calcium-signaling components in plants. Phytochemistry 65, 1745–1776 [DOI] [PubMed] [Google Scholar]
- 69. Michaels S. D. (2009) Flowering time regulation produces much fruit. Curr. Opin. Plant Biol. 12, 75–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kotake T., Takada S., Nakahigashi K., Ohto M., Goto K. (2003) Arabidopsis TERMINAL FLOWER 2 gene encodes a heterochromatin protein 1 homolog and represses both FLOWERING LOCUS T to regulate flowering time and several floral homeotic genes. Plant Cell Physiol. 44, 555–564 [DOI] [PubMed] [Google Scholar]
- 71. Giraudat J., Hauge B. M., Valon C., Smalle J., Parcy F., Goodman H. M. (1992) Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4, 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ulmasov T., Hagen G., Guilfoyle T. J. (1997) ARF1, a transcription factor that binds to auxin response elements. Science 276, 1865–1868 [DOI] [PubMed] [Google Scholar]
- 73. Kagaya Y., Ohmiya K., Hattori T. (1999) RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res. 27, 470–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Love J., Dodd A. N., Webb A. A. (2004) Circadian and diurnal calcium oscillations encode photoperiodic information in Arabidopsis. Plant Cell 16, 956–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Imaizumi T., Schroeder J. I., Kay S. A. (2007) In SYNC. The ins and outs of circadian oscillations in calcium. Sci. STKE 2007, pe32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.