Background: eIF2B is a multisubunit protein that regulates protein synthesis.
Results: We identify conserved residues in eIF2Bγ and eIF2Bϵ that are involved in interactions between subunits of the eIF2B complex.
Conclusion: Highly conserved regions and residues in eIF2B are crucial for creating eIF2B protein complexes.
Significance: This study provides new insights into the interactions that hold together the eIF2B holocomplex.
Keywords: Protein Domains, Protein Self-assembly, Protein Synthesis, Translation, Translation Initiation Factors, I-patch, eIF2B
Abstract
Eukaryotic initiation factor 2B (eIF2B) plays a key role in protein synthesis and in its control. It comprises five different subunits, α-ϵ, of which eIF2Bϵ contains the catalytic domain. Formation of the complete complex is crucial for full activity and proper control of eIF2B. Mutations in the genes for eIF2B cause an often severe neurological disorder, “vanishing white matter.” eIF2Bγ and eIF2Bϵ contain homologous and conserved domains with sequence similarity to nucleotidyl transferases (NTs) and acyl transferases and can form a binary complex. The latter contain a hexad repeat that mainly comprises isoleucyl residues (hence termed the “I-patch” region). These data reveal that certain residues in the NT domains of eIF2Bγ/ϵ, which are highly conserved throughout eukaryotes, play key roles in the interactions between subunits in the eIF2B complex. Our data show that the I-patch regions are important in the interactions between the catalytic eIF2Bγϵ complex and the other subunits. We also studied the functional effects of vanishing white matter mutations in the NT and I-patch domains. Lastly, our data show that eIF2Bγ promotes the expression of eIF2Bϵ, providing a mechanism for achieving correct stoichiometry of these eIF2B subunits in the cell.
Introduction
Eukaryotic initiation factor 2B1 (eIF2B)3 plays a key role in protein synthesis (mRNA translation) and its regulation (1). It acts as a GDP dissociation stimulator protein to mediate guanine nucleotide exchange on eIF2 (2), the factor that (as eIF2.GTP) brings the initiator methionyl-tRNA (Met-tRNAiMet) to the ribosome to recognize the start codon during each “round” of translation initiation. eIF2B is thus often termed a guanine-nucleotide exchange factor (GEF). The observations that overexpressing eIF2B increases protein synthesis rates in HEK293 cells (3) and cardiomyocytes (4) and induces growth of the latter indicate that eIF2B is a critical rate-determining component of the protein synthesis machinery. Consistent with this, the activity of eIF2B can be regulated in several different ways.
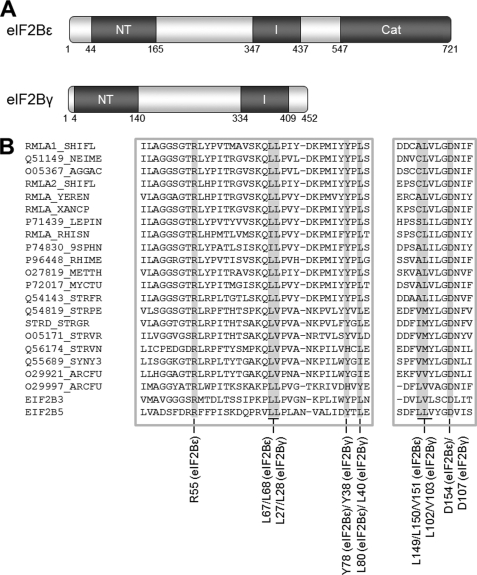
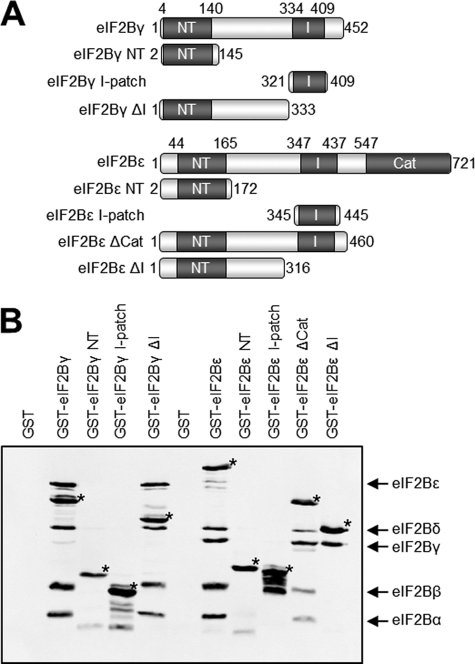
eIF2B comprises five non-identical subunits, α-ϵ, named by increasing size. eIF2Bϵ contains within its C-terminal region the catalytic domain (approximately residues 527–726 in human eIF2Bϵ), which mediates GDP/GTP exchange on eIF2 (5, 6) (Fig. 1A). The extreme C terminus interacts with eIF2 (7). Large sections of the sequences of eIF2Bγ and ϵ show mutual sequence similarity (8) (see Fig. 1A) and form a binary complex termed the “catalytic subcomplex.” Both of these subunits contain regions that show similarity to nucleotidyl transferases (NT) (residues 44–165 of eIF2Bϵ and residues 4–140 of eIF2Bγ, Fig. 1B) and to acyl transferases (AT) (residues 347–437 of human eIF2Bϵ and residues 334–409 of eIF2Bγ), although the functional significance of these resemblances is unclear. The latter region contains repeats with an approximate hexad spacing of hydrophobic branched chain amino acids (generally Ile but sometimes Val or Leu). In other proteins, such so-called “I-patches” form left-handed β-helices that may be involved in oligomerization (9, 10). Although these homologous regions were first reported as long ago as 1995 (8), their significance for the function of eIF2B has remained unclear.
FIGURE 1.
Conserved domains in eIF2Bγ and ϵ. A, the domain structure of eIF2Bγ and ϵ showing the conserved NT and I-patch (I) homology regions and the catalytic domain (Cat) with approximate residue numbering. B, an alignment of the NT homology regions of human eIF2Bγ and ϵ (bottom) alongside sequences from a number of proteins containing NT domains. The reference sequences used for alignment were NP_003898.2 and NP_065098.1. The figure was generated using Clustal W2 and Jalview software. Mutated amino acids are identified and are indicated with a gray background in the sequence alignment.
The other three subunits, α, β, and δ, also show mutual sequence similarity (11, 12) and form a “regulatory” subcomplex (13), so named because it confers on the holocomplex sensitivity to inhibition by the substrate eIF2 (14) when the latter is phosphorylated at Ser-51 on its α subunit. This provides one important mechanism for regulating eIF2B activity. In the case of mammalian eIF2B, the α subunit is required for full catalytic activity (2, 15, 16). It is also essential to confer sensitivity to phosphorylated eIF2 (17). Furthermore, although eIF2Bγϵ “catalytic” subcomplexes do show activity, this is markedly enhanced by the presence of the other subunits (16). Thus, costoichiometric expression of the subunits is necessary for the optimal activity and proper regulation of eIF2B. However, it is not known how this is achieved physiologically.
The importance of eIF2B in normal cell function is highlighted by its involvement in an often severe inherited neurodegenerative disease, known as leukoencephalopathy with vanishing white matter (VWM) (also termed CACH, childhood ataxia with central nervous system hypomyelination (18)). Patients with this disease show progressive demyelination of the white matter in the brain, leading to ataxia, mental retardation, and eventual death. A wide range of phenotypes has been observed, from very mild adult-onset forms to severe neonatal disease. VWM is an autosomal recessive disease requiring inherited mutations in both copies of the genes encoding the same subunit of eIF2B. Although mutations in the EIF2B5 gene, encoding the ϵ subunit, have been described most frequently, numerous VWM-causing variants have been identified in the genes encoding the remaining four eIF2B subunits (18, 19). Previous studies have postulated that a decrease in eIF2B activity caused by the mutations is responsible for the phenotype (reviewed in Ref. 18) and that the VWM phenotype is correlated with eIF2B activity (20, 21). However, we have recently demonstrated that this is not the case, and in fact, some of the most severe VWM mutations have barely any effect on eIF2B activity (16). Thus, the means by which mutations in the EIF2B1–5 genes cause VWM is not yet understood.
As alluded to above, cellular eIF2B activity is regulated by phosphorylation of eIF2 on Ser-51 in its α subunit, which converts eIF2 to a competitive inhibitor of eIF2B (22, 23). Four eIF2 kinases are known to exist in mammals. They are activated under different, usually stress-like conditions, including endoplasmic reticulum stress caused by an accumulation of unfolded or misfolded proteins and nutrient stress, in particular amino acid deprivation. Inhibition of eIF2B by phosphorylated eIF2 is dependent on eIF2Bα, the structure of which has been solved and contains a potential binding pocket for the phospho-serine group (24). Recombinant eIF2B complexes lacking the α subunit are not inhibited by phospho-eIF2 (17).
To date, there is limited information regarding the functional significance of the conserved domains in eIF2Bγ and ϵ. This is important for understanding both the assembly and function of the eIF2B complex and the roles that these domains play in mediating eIF2B function. The data in this report reveal that key residues within these conserved regions are crucial for the assembly of the holocomplex or its association with the regulatory subcomplex. We have also identified a role for eIF2Bγ in mediating the stability of eIF2Bϵ, a potential means of regulating the cellular levels of this subunit and, thus, eIF2B activity.
EXPERIMENTAL PROCEDURES
Plasmids
Plasmids encoding myc-tagged eIF2B subunits and His-myc-tagged eIF2Bγ and ϵ have been described previously (3, 25). We mutated specific residues in the His-myc-tagged subunits by site-directed mutagenesis as described previously (16). PCR fragments of NT and I-patch domains as well as truncated forms of eIF2Bγ and ϵ were cloned into pEBG-6P to produce GST-myc-tagged versions of these fragments.
Cell Culture and Lysate Preparation
HEK 293 cells were cultured and maintained as described previously (3). Cells were transfected by the calcium phosphate method and harvested after 48 h by washing twice in PBS and lysing the cells in lysis buffer (20 mm HEPES-KOH (pH 7.6), 50 mm β-glycerophosphate, 50 mm KCl, 0.5% Triton X-100, 0.5 mm NaVO3, 14.3 mm β-mercaptoethanol, 1× complete protease inhibitors (Roche)). Lysates were cleared by centrifugation at 20,000 × g at 4 °C for 15 min and stored at −80 °C.
Affinity Chromatography
Crude lysates from transfected cells were analyzed by SDS-PAGE/Western blot analysis (using an anti-myc tag antibody) (9E10, Sigma-Aldrich, Poole, UK). The amount of lysate used for each pull-down was carefully adjusted to ensure that equal amounts of the His- or GST-tagged subunits were used for each batch of experiments. Appropriate volumes of lysate were incubated with either nickel-nitriloacetic acid-agarose in the presence of 20 mm imidazole (for His-tagged subunits) or glutathione-Sepharose (for GST-tagged subunits) for 1 h at 4 °C. Complexes were washed with lysis buffer containing 10% (v/v) glycerol, 0.15% (v/v) Triton X-100, and 20 mm imidazole (for His6 pull-downs). The purified complexes were either analyzed by Western blot analysis or used to assay eIF2B activity (as described earlier) (16, 26).
Assays for eIF2B Activity
eIF2B (GEF) activity assays were carried out as described previously (25, 27) by measuring the release of [3H]GDP from eIF2 preparations by affinity-purified eIF2B complexes. Each experiment was carried out in triplicate on at least three separate eIF2B preparations. Data are expressed as mean ± S.E. (n ≥ 3), with the activity of complexes prepared with wild-type-tagged subunit normalized to 1.
Homology Modeling
Structural models of eIF2Bγ and ϵ were produced using the SWISS-MODEL workspace using the protein sequences of eIF2Bγ (NP_065098) and eIF2Bϵ (NP_003898). The locations of the mutated residues were examined in all the models produced to provide the best consensus of location.
RESULTS
Homology Modeling of eIF2Bγ and eIF2Bϵ
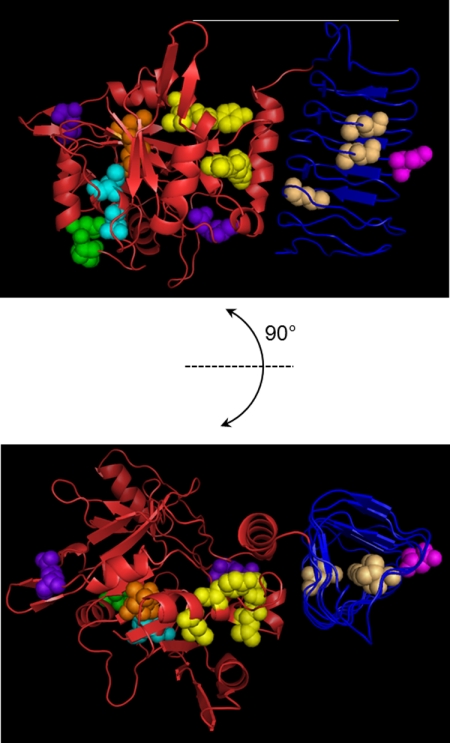
Because a number of proteins with a combination of NT and I-patch domains have been identified, and structures were solved for a number of these, we employed homology modeling to assist in identifying the location, potential roles, or mutated residues using the SWISS-MODEL server. This produced a number of partial models of eIF2Bγ and eIF2Bϵ that were used in combination to identify the locations of the mutated residues. We compared the locations of mutated residues on several models to increase the confidence in their position. A model of the region of eIF2Bϵ comprising the NT and I-patch domains, based on the structure of glucose-1-phosphate thymidyltransferase from Sulfolobus tokodaii (PDB code 2GG0), is shown in Fig. 2 as a representative example of the modeling output. None of the models reproduced the complete eIF2Bϵ structure, but information from the partial structures suggests that the I-patch region (Fig. 2, blue backbone) forms a separate domain to the polypeptide sequence prior to the I-patch (Fig. 2, red backbone).
FIGURE 2.
Homology model of structure of residues 43–450 of eIF2Bϵ. This model is on the basis of PDB code 2GGO showing the predicted structure of the NT domain and interdomain region (red backbone) and the I-patch domain (blue backbone) plus the location of key mutations in this study. Locations of mutations in the NT domain affecting binding to the α, β, and δ subunits are yellow. Mutated residues affecting binding to all subunits are shown in orange, and those affecting activity without affecting complex formation are shown in light blue and green (NF motif). Mutations with no effect on complex formation or eIF2B activity are shown in purple. The location of mutated residues in the I-patch domain affecting complex formation and/or activity are shown in brown, with the D387G VWM-associated mutation shown in pink. The model shows that the I-patch domain (blue backbone) forms a distinct domain from the NT and interdomain region (red backbone). The catalytic domain is not shown on this model.
Mutation of Residues in the eIF2Bϵ NT Domain
The NT homology region of human eIF2Bϵ encompasses (approximately) residues 44–165 (Fig. 1A). To assess the importance of individual residues in the function or assembly of the eIF2B complex, we mutated the residue(s) of interest in the cDNA of the relevant subunit (e.g. eIF2Bϵ) and expressed the mutant (with myc and His6 tags) in HEK293 cells, along with myc-tagged versions of the other subunits (in that case, eIF2Bα-δ). Following lysis, we purified the complexes on nickel- nitrilotriacetic-agarose (as described under “Experimental Procedures”) and analyzed the isolated complexes for eIF2B activity and eIF2B complex formation as described earlier (25). One caveat with this kind of approach is that mutations may affect the folding of the polypeptide. We therefore chose mainly to make mutations to a residue smaller than the original one, usually alanine, although we cannot rule out that this may still affect protein folding. Biophysical studies to confirm the correct folding of the many mutated proteins used here are, unfortunately, beyond the scope of this study.
In principle, the isolated complexes might also contain endogenous eIF2B subunits. However, we recently showed, using an approach identical to the one used here, that the levels of endogenous subunits that copurify with the recombinant eIF2B polypeptides is very low (indeed, they were undetectable (16)). Furthermore, for the studies using point mutants, the mutated subunit is always the one containing the hexahistidine tag used for purification so that the possibility of complexes containing a mixture of wild-type and ectopically expressed mutant eIF2B subunits cannot arise.
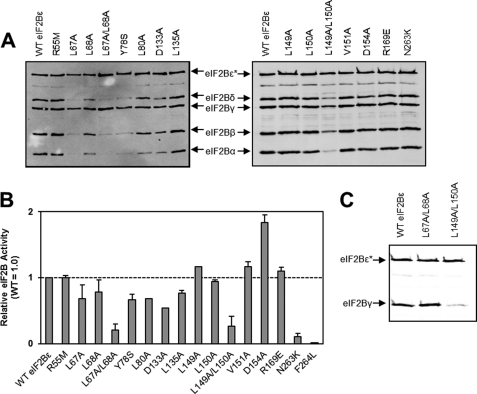
To assess whether the homology of this region to NTs is of significance for the catalytic function of eIF2B, we mutated a number of residues that are strongly conserved (Fig. 1B). The more N-terminal part of the NT-domain contains several residues that are present in all known eIF2Bϵ and eIF2Bγ sequences. This includes a group of adjacent, highly conserved residues (LLPL, residues 67–70 in human eIF2Bϵ, Fig. 1B). Interestingly, one of the residues within this motif is mutated in patients with VWM, L68S (28). Mutation of Leu-67 to alanine decreased the association of eIF2Bϵ with the α, β, and δ subunits and led to a 30% decrease in activity (Fig. 3, A and B). This decrease in activity is in line with the observation that human eIF2Bγϵ catalytic subcomplexes show only half the activity of the whole complex (16). In contrast, mutation of Leu-68 to alanine had no effect on complex formation and caused a 30% decrease in eIF2B activity, an effect identical to that we have described previously for the L68S VWM mutation (16). Mutation of both Leu-67 and Leu-68 to alanines led to loss of association with the α, β, and δ subunits and a severe decrease in eIF2B activity (Fig. 3, A and B). The loss of activity likely reflects the decreased association of these eIF2Bϵ mutants with eIF2Bα, β and δ, which is required for full activity of mammalian eIF2B (16). These residues are therefore critical for the interaction between the regulatory and catalytic subcomplexes and the formation of fully active eIF2B complexes.
FIGURE 3.
Analysis of the effects of mutations in the NT homology region of eIF2Bϵ. HEK293 cells were transfected with vectors encoding His/myc-tagged WT eIF2Bϵ or the indicated mutants plus myc-tagged versions of the other four eIF2B subunits (A) or wild-type myc-eIF2Bγ only (C). After analysis of the resulting lysates by SDS-PAGE Western blot analysis using anti-myc, samples containing similar amounts of His/myc-eIF2Bϵ were subjected to purification on Ni2+ beads to isolate recombinant eIF2Bϵ and associated subunits. Samples of the purified material were either analyzed by SDS-PAGE and Western blot analyses using anti-myc (A and C) or assayed for eIF2B (nucleotide exchange) activity (B). In A and C, the positions of the myc-tagged eIF2B subunits are shown. The asterisk indicates the His-tagged subunit (here, eIF2Bϵ).
Arg-55 in human eIF2Bϵ corresponds to a residue that is highly conserved among other NTs (8). Mutation of this residue to methionine (similar in size to arginine but uncharged) had no effect on either complex formation or eIF2B activity (Fig. 3, A and B).
Tyr-78 and Leu-80 (in eIF2Bϵ) are almost universally conserved in eIF2B sequences (the exception being the replacement of Tyr-78 to Phe, a very similar residue, in Caenorhabditis elegans). When Tyr-78 was mutated to a non-aromatic residue that still contains a hydroxyl group, serine, there was a strong impairment of association with the α, β, and δ subunits, and a decrease in eIF2B activity, similar to that observed for the L67A mutant (Fig. 3, A and B). Mutation of Leu-80 to alanine led to a modest effect on the interaction with eIF2Bα, and a decrease in activity similar to that seen for the Y78S mutant.
The C-terminal part of the NT domain contains an aspartate residue, Asp-154 in eIF2Bϵ, which is conserved in NTs and is predicted to be involved in Mg2+ binding (Fig. 1B, Ref. 8). Mutation of this residue to alanine, which lacks the negative charge required to bind Mg2+, had no effect on interactions with other subunits (Fig. 3, A and B). However, it surprisingly led to an apparent 2-fold increase in eIF2B activity. Although this residue is located outside of the core domain required for catalysis, it seems to play a role in eIF2B activity or its regulation.
Adjacent to Asp-154 lies a triad of branched-chain residues conserved by residue type in all known eIF2Bγ/ϵ and NT-domain sequences (LLV, residues 149–151, in human eIF2Bϵ; Fig. 1, A and B). Mutation of these residues individually to alanine had no significant effect on complex formation or eIF2B activity. However, mutation of both Leu-149 and Leu-150 to alanine caused a decrease in the association with the other four subunits, with a concomitant decrease in activity.
To confirm the effect of the L149A/L150A mutant in the interaction between eIF2Bγ and eIF2Bϵ, we transfected cells with His-myc-tagged wild-type, L67A/L68A, and L149A/L150A eIF2Bϵ with myc-tagged eIF2Bγ and no other subunits and then isolated the complexes as above. The data (Fig. 3C) show a clear decrease in the amount of eIF2Bγ associating with the L149A/L150A eIF2Bϵ but not the wild-type or L67A/L68A versions.
Asp-133 and Leu-135 are almost completely conserved among known eIF2Bγ and ϵ sequences (Fig. 1B). Mutation either of these residues to alanine led to a slight impairment of complex formation for the D133A mutant and a decrease in eIF2B activity for both mutants (Fig. 3, A and B). These mutants show greater deficits in activity than would be expected solely from effects on binding to other subunits (which is hardly affected by the L135A mutation, Fig. 3A), suggesting that these mutations may affect activity for additional reasons. Mutation of another completely conserved residue, Arg-169 in eIF2Bϵ, had no effect on either complex formation or activity (Fig. 3, A and B).
NT domain mutants have been observed in patients with VWM. As well as the L68S mutation described above, we have previously examined the effects of the VWM mutations F56V, V73G, T91A, R113H, and R136C on complex formation and eIF2B activity (16, 25). None of these mutations affected eIF2B complex formation, but several (T91A, R113H, and R136C) led to a decrease in eIF2B activity, whereas one, V73G, led to an increase.
These data demonstrate that the NT domain of eIF2Bϵ has an important role in the assembly of the eIF2B complex and, despite its remoteness from the core catalytic domain, in determining the activity of the eIF2B complex. In particular, we have identified several conserved residues that are critical for the interaction between eIF2Bϵ and the regulatory subcomplex, including Leu-67 and Tyr-78. Furthermore, we have demonstrated that mutation of Leu-149 and Leu-150 together is able to destabilize the interaction between eIF2Bγ and eIF2Bϵ.
The Asn-Phe Motif in eIF2Bϵ is Necessary for Activity in Human eIF2B
Gomez and Pavitt (5) showed that an Asn-Phe (NF) motif in yeast eIF2Bϵ (residues 249/250 in yeast, 263/264 in human) was important for facilitating the enhancement of eIF2B activity that results from the formation of holocomplexes. This motif is conserved in all available eIF2Bϵ sequences. We created mutations corresponding to those studied by Gomez and Pavitt (5), i.e. N263K and F264L. Both mutations had little effect on complex formation (Fig. 3A and data not shown) but showed a marked effect on eIF2B activity. The N263K mutation decreased activity by about 90%, whereas F264L was completely inactive (Fig. 3B).
Effects of Mutations of the eIF2Bγ NT Domain
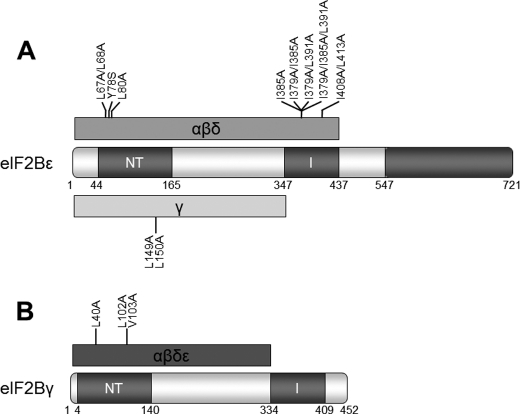
To compare the roles of conserved residues in the NT domain of eIF2Bϵ with the corresponding residues in eIF2Bγ, we mutated the relevant residues in eIF2Bγ that correspond to those in eIF2Bϵ whose mutation had the most severe effects on activity and/or complex formation (Fig. 4, A). In eIF2Bγ, the equivalent of the LLPL motif at 67–70 of eIF2Bϵ comprises residues 27–30, and the LVL motif at residues 149–151 in eIF2Bϵ is at 102–104 in eIF2Bγ (Fig. 1B). Unlike the corresponding residues in eIF2Bϵ, mutation of both Leu-27 and Leu-28 to alanines had little effect on complex formation, but it did have a similar effect on eIF2B activity (Fig. 4A,B). Mutation of both Leu-102 and Val-103 to alanine had a very similar effect to changing the equivalent residues in eIF2Bϵ, leading to a loss of interaction with all the other eIF2B subunits and, because eIF2Bϵ no longer copurifies efficiently with the eIF2Bγ(L102A/L103A) mutant, a substantial decrease in eIF2B activity (Fig. 4, A and B).
FIGURE 4.
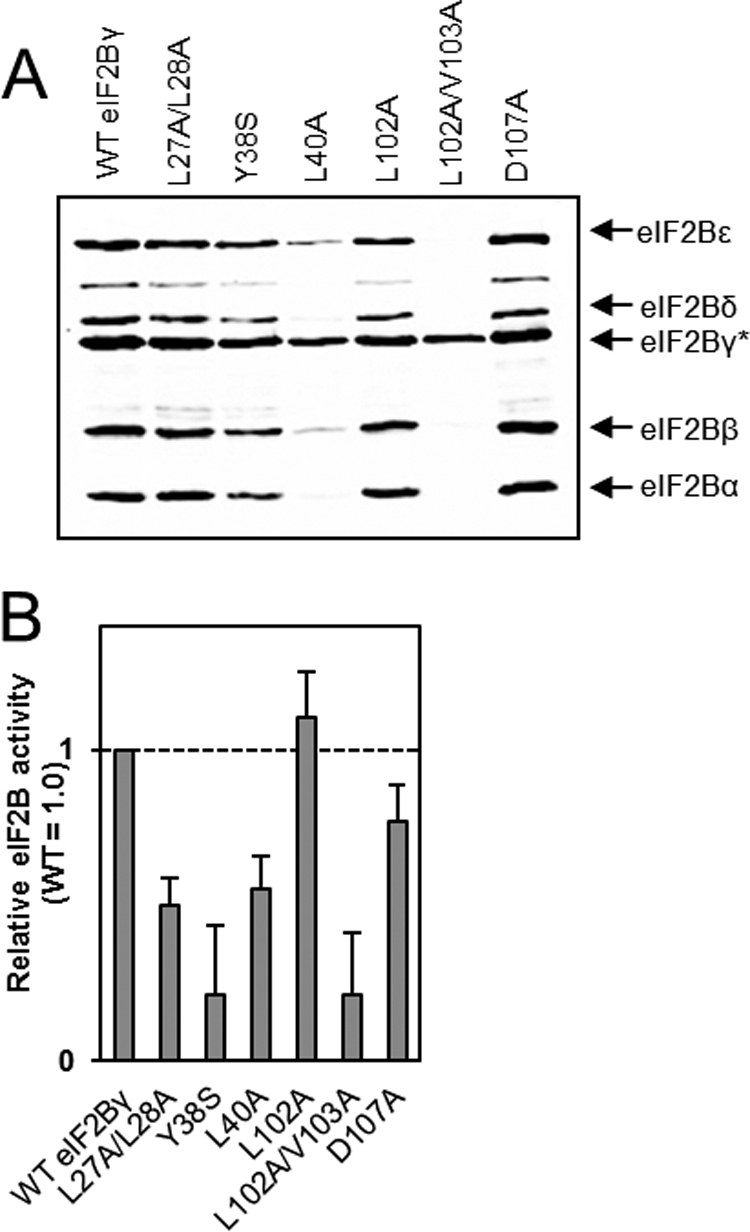
Analysis of the effects of mutations in the NT homology region of eIF2Bγ. HEK293 cells were transfected with vectors encoding His/myc-tagged WT eIF2Bγ or the indicated mutants plus myc-tagged versions of the other four eIF2B subunits. Lysates were analyzed as described in the legend to Fig. 2, except that lysates were normalized for expression of eIF2Bγ, and the asterisk here indicates that eIF2Bγ bore the His tag. A, analysis of purified recombinant eIF2B complexes by SDS-PAGE and Western blot analysis. B, activity data for complexes containing the indicated mutants of eIF2Bγ (using anti-myc).
Mutation of Tyr-38, the equivalent of Tyr-78 in eIF2Bϵ, had a less dramatic effect on complex formation, unlike in eIF2Bϵ, but had a severe effect on eIF2B activity (Fig. 4, A and B), suggesting that this residue affects catalytic function independently of an influence on the association of eIF2Bγ with the other subunits. Conversely, mutation of Leu-40, the equivalent of Leu-80 in eIF2Bϵ, to an alanine, which in eIF2Bϵ had only a minor effect on complex formation, affected interactions with all the other subunits in the complex, including eIF2Bϵ. Thus, less of the catalytic subunit copurifies with eIF2Bγ(L40A), likely explaining the substantial decrease in apparent GEF activity (Fig. 4, A and B).
Finally, mutation of Asp-107, which corresponds in position to Asp-154 in eIF2Bϵ, to alanine had no effect on complex formation, in a similar manner to its counterpart, but led to a slight decrease in activity rather than the increase observed for the D154A mutation in eIF2Bϵ (Fig. 4, A and B).
These data demonstrate that, although in some cases equivalent residues in eIF2Bγ and ϵ have similar roles in mediating intersubunit interactions and affecting eIF2B activity, this is not true in every case.
A Critical Role for the eIF2Bϵ I-patch Domain in the Interaction between the Catalytic and Regulatory Subcomplexes
The central part of eIF2Bϵ (residues 347–437) and the C-terminal part of eIF2Bγ (residues 366–409) show similarity to one another and members of the AT group of enzymes (8). The defining motif is a roughly hexad repeat of hydrophobic branched-chain amino acids, in particular isoleucine but also leucine and valine, often followed by a glycine. Similar features are known to form left- or right-handed β-helix structures, where the hydrophobic amino acids face inwards toward the center of the helix and stabilize the structure through hydrophobic interactions (29–33). The helices can associate with one another to stabilize protein structure or to form protein complexes, either through interactions between the “faces” of the helix or end-to-end (34). It seems therefore possible that the I-patch regions of eIF2Bγ and ϵ might serve an analogous function within the eIF2B complex.
To test this, we adopted a similar approach to that used for the NT domains and mutated residues conserved between eIF2Bγ and ϵ, particularly focusing on the repeating branched chain amino acids and analyzing their effects on complex formation and eIF2B activity. We also analyzed the effects of two VWM mutations in this region, I385V, which has been identified as a compound heterozygous mutation in patients with severe infantile disease (35) and D387G, which has been identified in patients with classical infantile disease, again as a compound heterozygous mutation (36).
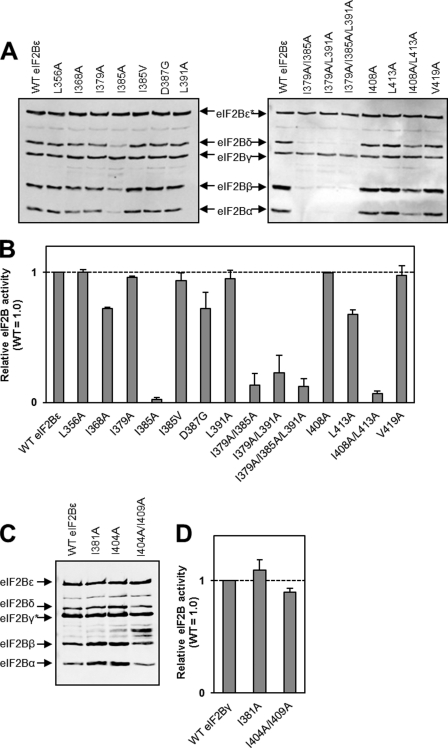
Mutation of the eIF2Bϵ residues Leu-356, Ile-379, Leu-391, Ile-408, and Val-419 individually to alanine residues had no effect on complex formation or eIF2B activity (Fig. 5A). Mutation of Ile-368 or Leu-413 to alanine led to an ∼30% decrease in eIF2B activity (Fig. 5B). Furthermore, the I368A mutant demonstrated a slight defect in interactions with the α, β, and δ subunits (Fig. 5A). Mutation of Ile-385 to alanine led to an almost complete loss of eIF2B activity, even though this mutant can still interact with eIF2Bγ (Fig. 5, A and B), presumably indicating that this mutation has a substantial effect on the structure of eIF2Bϵ. However, the VWM-associated mutation at this residue, I385V, which is a more conservative change (exchanging one branched-chain residue for another), had no effect on complex formation or eIF2B activity (Fig. 5, A and B). A second VWM mutation in this region, D387G, showed little effect on complex formation and a modest decrease in activity.
FIGURE 5.
Analysis of the effects of mutations in the I-patch region of eIF2Bϵ and eIF2Bγ. See the legend to Fig. 2 for details. A and C, analysis of purified recombinant eIF2B complexes by SDS-PAGE and Western blot analysis. B and D, activity data for complexes containing the indicated mutants of eIF2Bϵ/γ. The asterisk indicates the His-tagged subunit in each case. The vertical line in the right-hand part of A indicates that the figure contains two separate Western blot analyses, each of which contain wild-type control samples. The vertical lines in C indicate that the figure contains non-adjacent lanes from the same Western blot analysis.
Because the I385A mutation has such a severe effect on complex formation and activity, but the flanking I379A and L391A mutations do not, we created a double mutant, I379A/L391A. This mutant showed marked defects in complex formation and eIF2B activity (Fig. 5, A and B). A second double mutant, I408A/L413A, also showed a clear defect in complex assembly and eIF2B activity (Fig. 5, A and B). However, this mutant still copurified with eIF2Bγ and, to a limited extent, with the other subunits.
Given the importance of the I-patch region for the interaction of eIF2Bϵ with the regulatory subcomplex, we also tested a number of mutations within the corresponding region of eIF2Bγ. Interestingly, the effect of these mutations was not as severe as their counterparts in eIF2Bϵ. Mutation of Ile-381 to alanine, equivalent to the I385A mutant, had no effect on complex formation or activity (Fig. 5, C and D). The double mutation I404A/I409A, equivalent to I408A/L413A in eIF2Bϵ, did have a modest effect on interaction with the α, β, and δ subunits but caused only a very slight reduction in activity (Fig. 5, C and D).
The Individual NT and I-patch Domains Are Unable to Interact with Other eIF2B Subunits
Our mutagenesis data suggest that the NT domains of eIF2Bγ and ϵ are involved in the interaction between these two subunits, whereas both the NT and I-patch domains are required for interaction with the α, β, and δ subunits. To test this, we transfected cells with vectors encoding GST- and myc-tagged versions of the eIF2Bγ and ϵ NT and I-patch domains (Fig. 6A) together with vectors for the other eIF2B subunits. Because our homology model (Fig. 2) suggests that the NT homology region is part of a larger domain incorporating the sequence between the NT and I-patch homology regions, we created constructs that contain the NT-domain the sequence C-terminal to it and are thus truncated just prior to the I-patch (eIF2Bϵ ΔI and eIF2Bγ ΔI, Fig. 6A). Finally, because we have demonstrated previously that eIF2Bϵ with a 46-residue deletion within the catalytic domain is still able to interact fully with the other subunits (26), we also generated a construct encoding eIF2Bϵ truncated just after the I-patch domain (eIF2Bϵ ΔCat, Fig. 6A).
FIGURE 6.
Interactions between domains of eIF2B expressed in human cells. A, the individual I-patch and NT domains of eIF2Bγ and ϵ and fragments of both subunits truncated as shown were GST-myc tagged at their N termini. B, GST-tagged fragments were coexpressed in HEK293 cells with the remaining eIF2B subunits. Affinity purification using glutathione beads followed by Western blot analysis showed interactions between the eIF2B fragments and remaining subunits. GST-tagged subunits are marked with an asterisk and refer to the fragment identified in the lane header.
We subjected the lysates from cells transfected with combinations of these vectors to affinity purification on glutathione-Sepharose and analyzed the bound material for the presence of the other eIF2B subunits. Full-length GST-tagged eIF2Bγ and eIF2Bϵ were used as positive controls. The pull-downs showed that neither the NT nor the I-patch domains are able to interact stably with any of the other subunits on their own (Fig. 6B), whereas the full-length proteins can, as expected, associate with the rest of the complex.
eIF2Bϵ ΔCat was able to interact with all the other subunits (Fig. 6B), whereas eIF2Bϵ ΔI was only able to interact with eIF2Bγ. This supports our findings from the mutagenesis data, suggesting that both the NT and I-patch homology regions of eIF2Bϵ are required for the interaction between the regulatory and catalytic subcomplexes. These data also suggest that the entire domain comprising the NT-homology region and the sequence up to the start of the I-patch domain are required for the interaction with eIF2Bγ but that the I-patch domain is dispensable for this interaction.
Surprisingly, eIF2Bγ ΔI was able to copurify with all four of the other subunits as effectively as full-length eIF2Bγ did. This suggests that the eIF2Bγ I-patch domain is not required for interactions within the eIF2B complex (Fig. 6B).
To confirm these interactions, we coexpressed the truncated subunits with myc-hexahistidine-tagged versions of eIF2Bγ (for eIF2Bϵ truncations) or eIF2Bϵ (for eIF2Bγ truncations) and isolated complexes using nickel-nitrilotriacetic resin. These data (not shown) confirmed the interactions between the truncated subunits and the rest of the eIF2B complex.
eIF2Bγ Assists the Expression of eIF2Bϵ
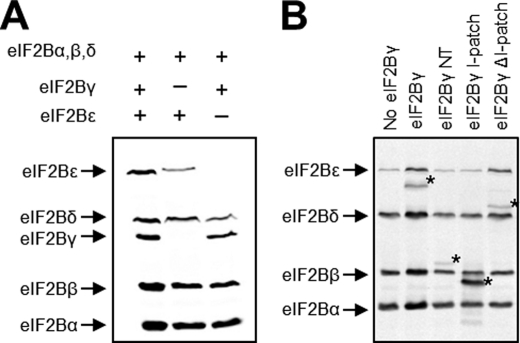
During Western blot analysis of cell lysates to equilibrate expression of His-tagged subunits prior to pull-down, we noted a decreased expression of ectopically expressed eIF2Bϵ in mutants that resulted in decreased interaction between eIF2Bγ and ϵ, regardless of whether the mutation was in eIF2Bγ or eIF2Bϵ. Therefore, we investigated whether expression of eIF2Bγ is required to stabilize expression of eIF2Bϵ. We transfected HEK293 cells with all five subunits or with all the subunits bar eIF2Bγ or eIF2Bϵ (Fig. 7A) and analyzed expression of eIF2Bϵ in the cell lysates by Western blotting. We found that omission of eIF2Bγ from the transfection lead to a 70% decrease in eIF2Bϵ expression compared with lysates expressing all five subunits (p < 0.01, n = 4). Furthermore, we observed a 35% decrease in expression of eIF2Bδ expression (p < 0.05). However, omission of eIF2Bϵ had no effect on expression of eIF2Bγ, but again resulted in a 35% decrease in eIF2Bδ expression.
FIGURE 7.
eIF2Bγ expression modulates the expression of other eIF2B subunits. A, Western blot analysis for myc-tagged eIF2B in lysates of cells expressing all five subunits or missing either eIF2Bγ or eIF2Bϵ. B, coexpression of fragments of eIF2Bγ with other eIF2B subunits. GST-tagged fragments are labeled with an asterisk and refer to the fragment identified in the lane header.
To investigate whether the interaction of eIF2Bγ with eIF2Bϵ was sufficient for stability of the latter, we expressed the eIF2Bγ NT and I-patch domains or eIF2Bγ ΔI. Western blotting of the lysates (Fig. 7B) showed that eIF2Bγ ΔI is able to stabilize ectopically expressed eIF2Bϵ but that the NT and I-patch domains alone are unable to stabilize eIF2Bϵ. Because eIF2Bγ ΔI is able to interact with eIF2Bϵ (Fig. 6B), unlike the NT and I-patch domains alone, we concluded that the interaction between eIF2Bγ and eIF2Bϵ was sufficient for stability of the latter.
DISCUSSION
To learn more about the structural organization of eIF2B, in particular the roles of the NT and I-patch domains, we analyzed the effects of a number of mutations of residues in these domains that are conserved, either between eIF2Bγ and ϵ, within eIF2B sequences from a large range of species, or conserved in the homologous domains, i.e. NT or I-patch domains. The key findings from these data demonstrate that both the NT and I-patch domains play key roles in the assembly of the eIF2B holocomplex (Fig. 8) and pinpoint the roles of highly conserved residues in the NT domain for interactions between eIF2Bγ and eIF2Bϵ. We have also identified residues in these domains that affect the activity of eIF2B, independently of effects on the ability to form complexes, despite being distant from the catalytic site. Furthermore, we show that expression of individual subunits, specifically eIF2Bδ and ϵ, is positively modulated by the presence of other subunits. Our data substantially extend earlier work that aimed to explore the regions of eIF2Bϵ that are involved in interactions with other subunits by expressing truncated versions of eIF2Bϵ in insect cells (37).
FIGURE 8.
Scheme indicating residues and domains involved in intersubunit interactions within eIF2B complexes. The conserved domains are indicated. Numbering is on the basis of the human protein sequences. A, domains and residues involved in interactions in eIF2Bϵ with other eIF2B subunits. The minimum region required for interaction with the “regulatory” subcomplex is shown in red, along with mutations affecting this interaction. The minimum region required for interaction with eIF2Bγ is shown in green, along with mutations identified as affecting this interaction. B, domains and residues involved in interactions of eIF2Bγ with other eIF2B subunits. The minimum region required for the interaction of eIF2Bγ with the other eIF2B subunits is shown in red, along with mutations of residues affecting these interactions.
The NT Homology Domain Mediates Subunit Interaction and Activity of eIF2B
A large number of proteins have been shown to contain domains homologous to nucleotidyl transferases (nucleoside diphosphate sugar pyrophosphorylases). The originally identified members of this family are involved in nucleoside diphosphate sugar formation in bacteria, but a number of other proteins have been identified with either an NT domain or with both NT and I-patch domains (8).
We investigated the role of this domain in the eIF2Bγ and ϵ subunits by mutating a number of residues that were either conserved within eIF2B sequences across a number of species or were conserved in eIF2B and NT sequences (Fig. 8). We identified several residues in conserved motifs in the NT domain, in particular the LLP (residues 67–69 of eIF2Bϵ and 27–29 of eIF2Bγ) and L(V/L)L (residues 149–151 of eIF2Bϵ and 101–103 of eIF2Bγ) motifs, as being critical for interactions with the regulatory subcomplex and, in the case of Leu-149/Leu-150 in eIF2Bϵ, for formation of the whole complex. Homology modeling of eIF2Bϵ suggests that Leu-67, Leu-68, Tyr-78, and Leu-80 lie on the surface of eIF2Bϵ, in a groove between the NT and I-patch domains (Fig. 7A, residues in yellow). These residues are thus likely either to directly mediate or to coordinate residues required for interaction with the subunits of the regulatory subcomplex. Leu-149 and Leu-150, however, are predicted to be buried within the interior of the domain (Fig. 7A, orange residues). Therefore, the observation that mutating them impairs binding of eIF2Bϵ to all the remaining subunits is likely to be due to destabilization of the domain structure, rather than reflecting a direct interaction at the interface between the subunits. This is consistent with the fact that mutating single residues in this region has no effect on subunit binding or eIF2B activity. The corresponding residues in eIF2Bγ and Leu-102/Val-103 are predicted to have a similar location in that protein, and again mutations of these residues led to decreased binding to the other subunits (Fig. 3A).
The other residues that we mutated, which had little or no effect on subunit association, are predicted to be present on the opposite face of the domain. Interestingly, the mutations that showed a defect in eIF2B activity, D133A and L135A (Fig. 7A, cyan residues), were modeled to be very close to the Asn-Phe motif at residues 263–264 (green residues), mutation of either of which has a dramatic effect on eIF2B activity but not on eIF2B complex formation. However, Arg-55 and Arg-169, mutations of which have no effect on complex formation or activity, are distant both from the residues affecting interactions and activity (Fig. 7A, purple residues).
Of particular interest was the effect of mutation of Asp-154 in eIF2Bϵ, which led to a 2-fold increase in eIF2B activity. This residue is predicted to bind a Mg2+ ion in nucleotidyl transferases that contributes to their enzyme activity. eIF2B has been reported to bind and be regulated by NAD(P)H/NAD(P)+ (27, 38). The present data could perhaps reflect an effect of the D154A mutation on nicotinamide adenine dinucleotide binding, given that that eIF2Bϵ does contain a region with homology to nucleotide-binding enzymes (a decrease in NAD(P)+ binding might relieve the reported inhibition by these compounds). However, it is not clear whether the ϵ subunit actually binds these nucleotides. Indeed, although several subunits of eIF2B do undergo photoaffinity labeling by 8-azido analogues of GTP or ATP, the labeled subunits did not include eIF2Bϵ but rather eIF2Bβ (8-azido-GTP) and probably eIF2Bγ/δ (8-azido-ATP)) (39). Further work is therefore needed to understand the basis of the effect of the D154A mutation.
The I-patch Domain of eIF2Bϵ Is Involved in Holocomplex Assembly
The functional significance of the I-patch domains of eIF2Bγ and ϵ, first noted in 1995 (8), has also not been studied previously. These domains contain a signature repeat of hydrophobic branched chain amino acids at approximately six residue intervals. This I-patch repeat is conserved across all known eIF2Bγ and eIF2Bϵ sequences, which is strongly suggestive of a crucial role in the structure and/or function of the eIF2B holocomplex. In other proteins, such domains have been shown to form β-helical structures stabilized by the repeating hydrophobic residues and have been shown to mediate protein-protein interactions within protein complexes (29–33). We therefore explored the possibility that these domains might be responsible for interactions within the catalytic subcomplex (because both eIF2Bγ and eIF2Bϵ contain this type of repeat). To test this, we mutated a number of the repeating residues to assess their functional significance.
A typical β-helix consists of three “faces” of β-strand structure forming a left-handed helix (34). This structure is stabilized by the hydrophobic branched chain amino acids facing into the core of the helix. Only two mutations of single residues (I368A and I385A) affected the association of eIF2Bϵ with other subunits. These residues are predicted to lie on the same face of the β-helix (Fig. 2, residues depicted in light brown). Mutation of these led to a decrease in both subunit interactions and eIF2B activity. The I385A mutation had the most dramatic effect on eIF2B activity of all the mutations in this study, virtually wiping out all eIF2B activity and seriously affecting binding to the regulatory subcomplex but not to eIF2Bγ. This activity is less than expected for eIF2Bγϵ binary complexes, which show 20% of the activity of the complete complex (16). Thus, these residues may have a direct influence on the activity of eIF2B. However, it is possible that this mutation destabilizes the I-patch domain and, therefore, affects the adjacent catalytic domain. This would not affect the interaction with eIF2Bγ because only the region of the subunit prior to the I-patch is required for this interaction. Curiously, a VWM-associated mutation at this residue, I385V, which is associated with severe infantile disease, albeit in a compound heterozygous manner with Y343C (36), had virtually no effect on eIF2B activity or complex formation. In contrast to the I385A mutation, the replacement of isoleucine by valine is relatively conservative. Furthermore, we found that mutation of the equivalent residue in eIF2Bγ (Ile-381) failed to mimic the effect on either complex formation or eIF2B activity. As we have reported previously, a number of VWM-associated mutations have no noticeable defect in either eIF2B activity, measured by GTP exchange on purified eIF2, or on complex formation (16).
Subunit Interactions Mediated by Individual Domains
A key finding for mutations in the I-patch domains of eIF2Bγ and ϵ was their lack of effect on the interaction between these two subunits, whereas certain mutations of the NT domain did affect this. To explore this further, we produced GST-tagged constructs of the NT and I-patch domains of eIF2Bγ and ϵ to investigate whether the individual domains alone were sufficient to interact with the other subunits. However, we found that, although they could be expressed in human cells, neither domain was able to interact with any of the other subunits. In the homology structure, the NT-domain does not appear to form a distinct domain but is part of a larger domain separate from the I-patch (Fig. 2, red backbone).Therefore, we produced constructs of eIF2Bγ and ϵ truncated prior to the I-patch domain (Fig. 7A). We found that eIF2Bϵ ΔI is only able to interact with eIF2Bγ (Fig. 7B). This confirmed our mutagenesis data that suggested that the eIF2Bϵ I-patch domain is not necessary for the interaction between eIF2Bγ and ϵ. In contrast, eIF2Bγ ΔI was able to interact with all the other subunits (Fig. 7B). This suggests that the eIF2Bγ I-patch is unnecessary for interactions within the eIF2B holocomplex (Fig. 8).
We have demonstrated previously that eIF2Bϵ with a 46-residue deletion in the catalytic domain is still able to interact with the remaining subunits (26). Therefore, we produced a truncation of eIF2Bϵ just after the I-patch domain (Fig. 7A). This truncated version contains the entire region of eIF2Bϵ homologous to eIF2Bγ. As expected, this truncated version of eIF2Bϵ was able to interact with the other subunits (Fig. 7B), suggesting that the regions of eIF2Bϵ not homologous to eIF2Bγ are not required for intersubunit interactions.
Mutual Facilitation of Expression of eIF2B Subunits
Expression of a multi-subunit complex within a cell requires tight regulation to ensure that equal quantities of each subunit are produced. This regulation can occur at the level of transcription, translation, or protein stability and may encompass all these mechanisms. We noticed that the expression level of eIF2Bϵ was always decreased in lysates where mutated subunits that affected interactions within the catalytic subcomplex were used, regardless of whether the mutated subunit was eIF2Bγ or ϵ. We investigated this further by overexpressing all five myc-tagged eIF2B subunits and comparing this to cells where the subunits were expressed, except eIF2Bγ or eIF2Bϵ. We found that omitting eIF2Bγ led to a significant (70%) decrease in expression of ectopic eIF2Bϵ, whereas omitting eIF2Bϵ had no effect on eIF2Bγ expression (Fig. 7A). Furthermore, absence of either subunit led to a decrease in eIF2Bδ expression. We saw no effect on expression of the α or β subunits (Fig. 7A).
Because our expression constructs do not contain the native untranslated regions of any of the subunits, we hypothesized that interaction between eIF2Bγ and ϵ was the stabilizing factor. Therefore, we expressed truncations and individual domains of eIF2Bγ (Fig. 7A) to see whether these were able to stabilize the ectopically expressed eIF2Bϵ. We found that only eIF2Bγ ΔI, the only truncated polypeptide which can interact with eIF2Bϵ, is able to restabilize ectopic eIF2Bϵ (Fig. 7B).
This enhancement of eIF2Bϵ expression by eIF2Bγ may provide a novel form of regulation of eIF2B, whereby expression of eIF2Bϵ, the catalytic subunit, is modulated by expression of the other subunits. This could allow tight control over the levels of eIF2Bϵ and, consequently, eIF2B activity. Such regulation may be very important because increased eIF2B activity can increase the rate of cell growth (4) and so may be a mechanism of tumorigenesis.
The interactions within a multisubunit complex such as eIF2B are crucial in defining its function and stabilization. Such interactions are frequently mediated by conserved domain structures and, particularly, conserved residues in such domains. Here we have examined the role of a number of residues in two such domains, the NT and acyl transferase (I-patch) domains of the γ and ϵ subunits of eIF2B, which comprise the catalytic subcomplex. We have shown that these domains play a role in mediating the interactions between the catalytic and regulatory subcomplex (containing the α, β, and δ subunits of eIF2B) as well as interactions within the subcomplex. We have also shown that a number of these residues are important for full activity of eIF2B and may be involved in external regulatory mechanisms of eIF2B.
Quite independently of this study, Reid et al. (40) adopted a different but complementary approach to investigate the interactions between subunits of the yeast eIF2B complex in which they expressed domains of eIF2B and tested their abilities to interact with other eIF2B subunits or their domains. Their data are generally in good agreement with ours, although in some cases the domains used in the two studies are not equivalent, making direct comparison more difficult. Their data also show that the conserved aspartate in the NT domain, which is Asp-154 in the human protein, is also dispensable for GEF function.
Interestingly, we have also identified a potential new means by which the cellular levels of eIF2B can be regulated through the eIF2Bγ-mediated augmentation of the expression of eIF2Bϵ, the catalytic subunit. This mechanism could operate to ensure costoichiometric expression of eIF2Bγ and eIF2Bϵ, the key components of the catalytic subcomplex. Maintaining correct levels of eIF2B is probably a critical parameter governing rates of protein synthesis, given its central role in regulating translation initiation (1) and the fact it is the least abundant of the main translation initiation factors (26).
Acknowledgments
We thank Hua Tang and Dr. Yanni Wang for skilled assistance. We thank Graham Pavitt (Manchester) for useful discussions and sharing data prior to submission of his laboratory findings.
Footnotes
- eIF2B
- eukaryotic factor 2B
- GEF
- guanine nucleotide exchange factor
- NT
- nucleotidyl transferase
- AT
- acyl transferase
- VWM
- vanishing white matter.
REFERENCES
- 1. Pavitt G. D. (2005) eIF2B, a mediator of general and gene-specific translational control. Biochem. Soc. Trans. 33, 1487–1492 [DOI] [PubMed] [Google Scholar]
- 2. Williams D. D., Price N. T., Loughlin A. J., Proud C. G. (2001) Characterization of the mammalian initiation factor eIF2B complex as a GDP dissociation stimulator protein. J. Biol. Chem. 276, 24697–24703 [DOI] [PubMed] [Google Scholar]
- 3. Wang X., Proud C. G. (2008) A novel mechanism for the control of translation initiation by amino acids, mediated by phosphorylation of eukaryotic initiation factor 2B. Mol. Cell. Biol. 28, 1429–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hardt S. E., Tomita H., Katus H. A., Sadoshima J. (2004) Phosphorylation of eukaryotic translation initiation factor 2Bϵ by glycogen synthase kinase-3β regulates β-adrenergic cardiac myocyte hypertrophy. Circ. Res. 94, 926–935 [DOI] [PubMed] [Google Scholar]
- 5. Gomez E., Pavitt G. D. (2000) Identification of domains and residues within the epsilon subunit of eukaryotic translation initiation factor 2B (eIF2Bϵ) required for guanine nucleotide exchange reveals a novel activation function promoted by eIF2B complex formation. Mol. Cell. Biol. 20, 3965–3976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gomez E., Mohammad S. S., Pavitt G. D. (2002) Characterization of the minimal catalytic domain within eIF2B. The guanine-nucleotide exchange factor for translation initiation. EMBO J. 21, 5292–5301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Asano K., Krishnamoorthy T., Phan L., Pavitt G. D., Hinnebusch A. G. (1999) Conserved bipartite motifs in yeast eIF5 and eIF2Bϵ, GTPase-activating and GDP-GTP exchange factors in translation initiation, mediate binding to their common substrate eIF2. EMBO J. 18, 1673–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Koonin E. V. (1995) Multidomain organization of eukaryotic guanine nucleotide exchange translation initiation factor eIF-2B subunits revealed by analysis of conserved sequence motifs. Protein Sci. 4, 1608–1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Steinbacher S., Seckler R., Miller S., Steipe B., Huber R., Reinemer P. (1994) Crystal structure of P22 tailspike protein. Interdigitated subunits in a thermostable trimer. Science 265, 383–386 [DOI] [PubMed] [Google Scholar]
- 10. Yang Z., Zhou Y., Liu K., Cheng Y., Liu R., Chen G., Jia Z. (2003) Computational study on the function of water within a β-helix antifreeze protein dimer and in the process of ice-protein binding. Biophys. J. 85, 2599–2605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bushman J. L., Asuru A. I., Matts R. L., Hinnebusch A. G. (1993) Evidence that GCD6 and GCD7, translational regulators of GCN4, are subunits of the guanine nucleotide exchange factor for eIF-2 in Saccharomyces cerevisiae. Mol. Cell. Biol. 13, 1920–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Paddon C. J., Hannig E. M., Hinnebusch A. G. (1989) Amino acid sequence similarity between GCN3 and GCD2, positive and negative translational regulators of GCN4. Evidence for antagonism by competition. Genetics 122, 551–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pavitt G. D., Ramaiah K. V., Kimball S. R., Hinnebusch A. G. (1998) eIF2 independently binds two distinct eIF2B subcomplexes that catalyze and regulate guanine-nucleotide exchange. Genes Dev. 12, 514–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pavitt G. D., Yang W., Hinnebusch A. G. (1997) Homologous segments in three subunits of the guanine nucleotide exchange factor eIF2B mediate translational regulation by phosphorylation of eIF2. Mol. Cell. Biol. 17, 1298–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Craddock B. L., Proud C. G. (1996) The α subunit of the mammalian guanine nucleotide exchange factor eIF-2B is essential for catalytic activity in vitro. Biochem. Biophys. Res. Commun. 220, 843–847 [DOI] [PubMed] [Google Scholar]
- 16. Liu A. R., van der Lei H. D., Wang X., Wortham N. C., Tang H., Van Berkel C. G., Mufunde T. A., Huang W., Van der Knaap M. S., Scheper G. C., Proud C. G. (2011) Hum. Mutat. 32, 1036–1045 [DOI] [PubMed] [Google Scholar]
- 17. Fabian J. R., Kimball S. R., Heinzinger N. K., Jefferson L. S. (1997) Subunit assembly and guanine nucleotide exchange activity of eukaryotic initiation factor-2B expressed in Sf9 cells. J. Biol. Chem. 272, 12359–12365 [DOI] [PubMed] [Google Scholar]
- 18. Van der Knaap M. S., Bugiani M., Boor I., Proud C. G., Scheper G. C. (2010) The Online Metabolic and Molecular Bases of Inherited Diseases, Chap. 235.1, (Valle D., Beaudet A. L., Vogelstein B., Kinzler K. W., Antonarakis S. E., Ballabio A., Scriver C. R., Sly W. S., Childs B., eds) McGraw-Hill, New York [Google Scholar]
- 19. Pavitt G. D., Proud C. G. (2009) Protein synthesis and its control in neuronal cells with a focus on vanishing white matter disease. Biochem. Soc. Trans. 37, 1298–1310 [DOI] [PubMed] [Google Scholar]
- 20. Fogli A., Schiffmann R., Hugendubler L., Combes P., Bertini E., Rodriguez D., Kimball S. R., Boespflug-Tanguy O. (2004) Decreased guanine nucleotide exchange factor activity in eIF2B-mutated patients. Eur. J. Hum. Genet. 12, 561–566 [DOI] [PubMed] [Google Scholar]
- 21. Horzinski L., Huyghe A., Cardoso M. C., Gonthier C., Ouchchane L., Schiffmann R., Blanc P., Boespflug-Tanguy O., Fogli A. (2009) Eukaryotic initiation factor 2B (eIF2B) GEF activity as a diagnostic tool for EIF2B-related disorders. PLoS. ONE 4, e8318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rowlands A. G., Panniers R., Henshaw E. C. (1988) The catalytic mechanism of guanine nucleotide exchange factor action and competitive inhibition by phosphorylated eukaryotic initiation factor 2. J. Biol. Chem. 263, 5526–5533 [PubMed] [Google Scholar]
- 23. Wek R. C., Jiang H. Y., Anthony T. G. (2006) Coping with stress. eIF2 kinases and translational control. Biochem. Soc. Trans. 34, 7–11 [DOI] [PubMed] [Google Scholar]
- 24. Hiyama T. B., Ito T., Imataka H., Yokoyama S. (2009) Crystal structure of the α subunit of human translation initiation factor 2B. J. Mol. Biol. 392, 937–951 [DOI] [PubMed] [Google Scholar]
- 25. Li W., Wang X., Van Der Knaap M. S., Proud C. G. (2004) Mutations linked to leukoencephalopathy with vanishing white matter impair the function of the eukaryotic initiation factor 2B complex in diverse ways. Mol. Cell. Biol. 24, 3295–3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. (2011) Global quantification of mammalian gene expression control. Nature 473, 337–342 [DOI] [PubMed] [Google Scholar]
- 27. Oldfield S., Proud C. G. (1992) Purification, phosphorylation and control of the guanine-nucleotide-exchange factor from rabbit reticulocyte lysates. Eur. J. Biochem. 208, 73–81 [DOI] [PubMed] [Google Scholar]
- 28. Ohlenbusch A., Henneke M., Brockmann K., Goerg M., Hanefeld F., Kohlschutter A., Gartner J. (2005) Hum. Mutat. 25, 411. [DOI] [PubMed] [Google Scholar]
- 29. Yoder M. D., Keen N. T., Jurnak F. (1993) New domain motif. The structure of pectate lyase C, a secreted plant virulence factor. Science 260, 1503–1507 [DOI] [PubMed] [Google Scholar]
- 30. Yoder M. D., Jurnak F. (1995) Protein motifs. 3. The parallel β helix and other coiled folds. FASEB J. 9, 335–342 [DOI] [PubMed] [Google Scholar]
- 31. Pickersgill R., Jenkins J., Harris G., Nasser W., Robert-Baudouy J. (1994) The structure of Bacillus subtilis pectate lyase in complex with calcium. Nat. Struct. Biol. 1, 717–723 [DOI] [PubMed] [Google Scholar]
- 32. Raetz C. R., Roderick S. L. (1995) A left-handed parallel β helix in the structure of UDP-N-acetylglucosamine acyltransferase. Science 270, 997–1000 [DOI] [PubMed] [Google Scholar]
- 33. Kisker C., Schindelin H., Alber B. E., Ferry J. G., Rees D. C. (1996) A left-hand β helix revealed by the crystal structure of a carbonic anhydrase from the archaeon Methanosarcina thermophila. EMBO J. 15, 2323–2330 [PMC free article] [PubMed] [Google Scholar]
- 34. Schulz E. C., Ficner R. (2011) Knitting and snipping. Chaperones in β-helix folding. Curr. Opin. Struct. Biol. 21, 232–239 [DOI] [PubMed] [Google Scholar]
- 35. Fogli A., Dionisi-Vici C., Deodato F., Bartuli A., Boespflug-Tanguy O., Bertini E. (2002) A severe variant of childhood ataxia with central hypomyelination/vanishing white matter leukoencephalopathy related to EIF21B5 mutation. Neurology 59, 1966–1968 [DOI] [PubMed] [Google Scholar]
- 36. Fogli A., Schiffmann R., Bertini E., Ughetto S., Combes P., Eymard-Pierre E., Kaneski C. R., Pineda M., Troncoso M., Uziel G., Surtees R., Pugin D., Chaunu M. P., Rodriguez D., Boespflug-Tanguy O. (2004) The effect of genotype on the natural history of eIF2B-related leukodystrophies. Neurology 62, 1509–1517 [DOI] [PubMed] [Google Scholar]
- 37. Anthony T. G., Fabian J. R., Kimball S. R., Jefferson L. S. (2000) Identification of domains within the ϵ subunit of the translation initiation factor eIF2B that are necessary for guanine nucleotide exchange activity and eIF2B holoprotein formation. Biochim. Biophys. Acta 1492, 56–62 [DOI] [PubMed] [Google Scholar]
- 38. Dholakia J. N., Mueser T. C., Woodley C. L., Parkhurst L. J., Wahba A. J. (1986) The association of NADPH with the guanine nucleotide exchange factor from rabbit reticulocytes. A role of pyridine dinucleotides in eukaryotic polypeptide chain initiation. Proc. Natl. Acad. Sci. U.S.A. 83, 6746–6750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dholakia J. N., Francis B. R., Haley B. E., Wahba A. J. (1989) Photoaffinity labeling of the rabbit reticulocyte guanine nucleotide exchange factor and eukaryotic initiation factor 2 with 8-azidopurine nucleotides. Identification of GTP- and ATP-binding domains. J. Biol. Chem. 264, 20638–20642 [PubMed] [Google Scholar]
- 40. Reid P. J., Mohammad-Qureshi S. S., Pavitt G. D. (2012) Identification of intersubunit domain interactions within eukaryotic initiation factor (eIF) 2B, nucleotide exchange factor for translation initiation. J. Biol. Chem. 287, 8275–8285 [DOI] [PMC free article] [PubMed] [Google Scholar]