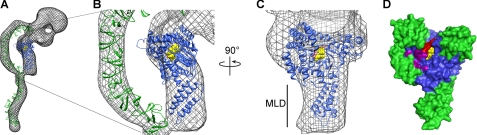
FIGURE 5.
Structure of the TcdA GTD alone and in the context of the TcdA holotoxin structure. A–C, three-dimensional reconstruction of TcdA (32) filtered to 25 Å is shown as a mesh surface with the crystal structure of the TcdA GTD (blue) and a model of the TcdA RBD (52) (green) placed into the density. UDP-glucose is shown as yellow spheres. In C, the model of the binding domain and the corresponding map density are removed. D, surface of the TcdA GTD shown in the same orientation as in C. The core GT-A fold is shown in blue with the additional α-helical regions in green (as in Fig. 1). The 516–522 loop is colored red, and UDP-glucose is represented as yellow spheres. Amino acids Lys-448, Gln-454, Glu-460, Arg-462, and Gly-471 are shown in purple. The corresponding residues in TcdB have been shown to be involved in substrate binding (45).