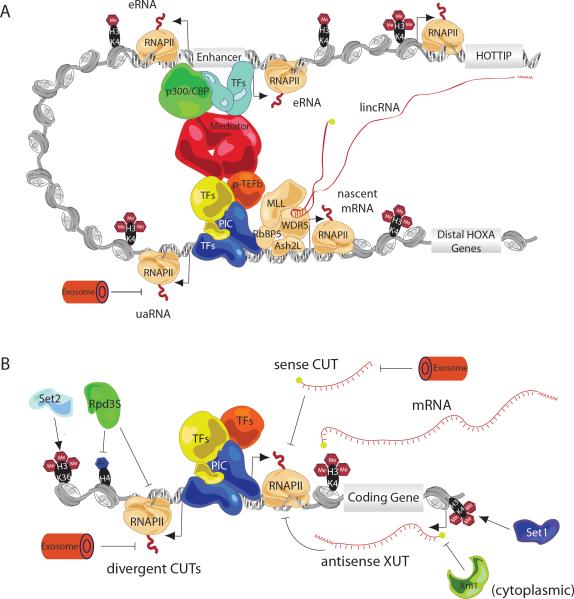
Figure 1. Schematic Diagram of Mammalian and Yeast lncRNA-chromatin Interactions.
A. Organization of a prototypical mammalian protein coding gene promoter, enhancer and lncRNA locus. The promoter of a protein-coding gene (e.g. distal HOXA genes) generates short sense and uaRNAs (degraded by the exosome) from divergently oriented sense and antisense RNAPII complexes. An enhancer element that regulates activity of the protein-coding gene forms a chromosomal loop to the promoter region through transcription factor and Mediator bridges. This enhancer generates eRNAs in a divergent manner marking it as a functional enhancer element. A lncRNA (HOTTIP) locus is brought in close proximity to protein-coding genes and the lncRNA transcript directly participates in targeting a chromatin modifying complex (MLL) to the promoter region to establish an active chromatin domain.
B. Organization of a prototypical yeast protein coding gene promoter and associated lncRNAs. The promoter of a protein-coding gene will produce sense and antisense (divergent) CUTs, which are substrates for the nuclear exosome. Sense CUTs are anti-correlated with mRNA levels. The upstream region of the promoter is often near the 3' end of another protein-coding gene and thus experiences Set2-mediated trimethylation of H3K36 and H4 deacetylaton by Rpd3S resulting in repression of the antisense CUT transcripts. Downstream of the promoter, antisense XUTs are activated by Set1 trimethylation of H3K4 and mediate mRNA repression. The XUT transcripts are degraded in the cytoplasm by the 5' to 3' exonuclease Xrn1.