Abstract
Aberrant RNA splicing is thought to play a key role in tumorigenesis. The assessment of its specific contributions is limited by the complexity of information derived from genome-wide array-based approaches. We describe how performing splicing factor-specific comparisons using both tumor and cell line datasets may more readily identify physiologically relevant tumor-specific splicing events. Affymetrix exon array data derived from glioblastoma (GBM) tumor samples with defined PTBP1 levels were compared with data from U251 GBM cells with and without PTBP1 knockdown. This comparison yielded overlapping gene sets that comprised only a minor fraction of each dataset. The identification of a novel GBM-specific splicing event involving the USP5 gene led us to further examine its role in tumorigenesis. In GBM, USP5 generates a shorter isoform 2 through recognition of a 5′ splice site within exon 15. Production of the USP5 isoform 2 was strongly correlated with PTBP1 expression in GBM tumor samples and cell lines. Splicing regulation was consistent with the presence of an intronic PTBP1 binding site and could be modulated through antisense targeting of the isoform 2 splice site to force expression of isoform 1 in GBM cells. The forced expression of USP5 isoform 1 in two GBM cell lines inhibited cell growth and migration, implying an important role for USP5 splicing in gliomagenesis. These results support a role for aberrant RNA splicing in tumorigenesis and suggest that changes in relatively few genes may be sufficient to drive the process.
Keywords: Exon Array, USP5, isopeptidase-T, GBM
INTRODUCTION
With a 2-year survival rate of less than 10%, glioblastoma multiforme (GBM) is among the most lethal cancers [1]. The high morbidity and mortality rates associated with GBM have led to the use of genomic approaches in multiple studies aimed at defining the underlying causes of this cancer. Indeed, GBM was among the first tumor types included for study as part of The Cancer Genome Atlas project [2]. From the broad application of genomic approaches has emerged a molecular classification of glioma subtypes and the beginnings of stratified treatment strategies based on genetic profiling (reviewed in [3]). However, gaps remain in our ability to use this wealth of acquired data as a tool for defining new treatment targets. With the continued development of methods for examining genome-wide RNA splicing has come a greater appreciation for the role of RNA processing in the creation of genetic diversity and regulation of cellular function. Results from recent studies using high-throughput sequencing approaches suggest that over 90% of protein-coding genes produce alternative mRNAs [4,5]. In 2006, Srebrow and Kornblihtt proposed a global pathway of RNA splicing disruption leading to tumorigenesis [6]. They recognized that tumor-specific changes in the composition or activity of the spliceosome could drive the alteration of RNA splicing patterns. One of the earliest studies of spliceosome gene expression in tumorigenesis, which included 145 genes involved in splicing, revealed widespread aberrant gene expression in brain, breast, colon, and prostate cancer [7]. More recently, we demonstrated that aberrant expression of genes involved in RNA processing could be used to segregate GBM samples from nontumor samples [8,9].
Although the overexpression of splicing factors has been found in various cancer types, whether it plays a direct role in cancer initiation and progression remains to be fully addressed. Two lines of evidence support the concept of a “cancer spliceosome”, which as a result of broad changes in splicing regulators would favor the production of cancer-specific mRNA isoforms. First, an ever-growing abundance of alternative mRNA isoforms has been found to be aberrantly expressed in tumors [6,9-11], and it is clear that the protein products derived many of these alternative mRNA isoforms would provide a selective growth advantage. For example, antiapoptotic isoforms of BCL2L1, CASP2, and FAS are often preferentially expressed in tumors [12]. Second, aberrant expression of at least four spliceosomal proteins, SFRS1, PTBP1, SF1, and SRp20, has been directly associated with tumorigenesis [13-16]. The overexpression of SFRS1 in NIH 3T3 cells is sufficient to allow anchorage-independent cell growth in soft agar and tumor formation in nude mice [15]. Targeted reduction of PTBP1, SF1, or SRp20 has been found to inhibit tumor cell growth and invasion [13,14,16]. However, aberrant splicing factor expression does not universally enhance tumor cell growth. Altering PTBP1 expression in cancer cell lines of various tissue origins caused both promotion and suppression of the malignant phenotype [17]. In glioma, we have demonstrated that PTBP1 expression is highly correlated with malignancy [18] and have defined specific roles for PTBP1 overexpression in cell proliferation and migration [19]. Through exon array analysis and functional studies, we also defined aberrant splicing of the RTN4 gene as a mediator of cell proliferation and migration [19]. Two recent studies have defined a PTBP1-regulated splicing event within the PKM2 gene that contributes GBM tumorigenesis [20,21]. These findings prompted a reevaluation of GBM array datasets with defined PTBP1 expression. This reevaluation did not identify PKM2 as a regulatory target of PTBP1, but did reveal USP5 as a novel target with a role in GBM cell migration.
MATERIALS AND METHODS
Array Data Sets
Exon array datasets for U251 cells and patient samples were previously generated in our laboratory [8,19]. Briefly, U251 cells were treated with either control or PTBP1 exon 2 morpholino oligonucleotide, and RNA was isolated and converted to cDNA and then to cRNA. The cRNA was labeled and hybridized to Affymetrix Human Exon Array 1.0 ST chips (Affymetrix, Santa Clara, CA). The exon array chip was scanned and a CEL file was created for each sample with GeneChip Operating Software. Each treatment was done in triplicate. A similar procedure was conducted for 12 nontumor brain samples and 24 grade IV GBM samples [8]. The 36 CEL files obtained from the patient samples were then clustered by PTBP1 expression levels using GeneSpring GX 11.0.1 software version 4 for Mac OS X (Agilent Technologies Inc., and Strand Life Sciences Pvt. Ltd., Santa Clara, CA). The studies presented here use only the 10 highest- and 10 lowest-expressing array samples.
Cell Culture and RNA Samples
The U251 and LN229 GBM cell lines were cultured using standard methods with Dulbecco’s modified Eagle’s medium (high glucose) supplemented with 10% fetal bovine serum at 37° C in 5% CO2. The collection of nontumor and GBM samples has been described previously [8]. Normal human astrocyte and cortical neuron RNA was purchased from ScienCell Research (Carlsbad, CA).
Qualitative and Quantitative Reverse-transcription Polymerase Chain Reaction (RT-PCR)
We performed RNA isolation and cDNA synthesis as described previously [8,19]. Qualitative PCR assays were performed for 32-35 cycles in a reaction consisting of cDNA, 10x PCR buffer, 50 mM MgCl2, 10 mM dNTPs, 20 ρmol of each of the forward and reverse primers, and Taq polymerase (5 units). All reagents in this reaction were purchased from Invitrogen (Carlsbad, CA). Specific cycling conditions were optimized for each primer pair and are available upon request. The resulting products were subjected to electrophoresis in a 1.2% agarose gel and visualized using ethidium bromide. Primer pairs were selected using Primer3 [22]. Primer sets included USP5 forward 5′-TGCCTCATTCCCTGACTACC-3′, USP5 reverse 5′-CACCAGCTGGATGATGACTG-3′. Primers used to test other genes are available on request. Quantitative RT-PCR was performed using both TaqMan and SYBR Green approaches with the StepOne Real-Time PCR system from Applied Biosystems (Foster City, CA). We determined PTBP1 expression levels with the TaqMan assay (Hs00259176-m1), which specifically measures the presence of exon 2. Ribosomal RNA (18S) (4308329) was used as an endogenous control to perform the comparative CT method. Both TaqMan assays were performed according to the manufacturer’s suggested protocol. To measure expression level of the USP5 isoform 1, we performed a SYBR Green assay using a forward primer designed to exclusively detect isoform 1 through hybridization to the alternative portion of exon 15 and a reverse primer in exon 16. We measured total USP5 levels with a primer set to detect USP5 exons 8-10 as the control to perform the comparative CT method. Thus, the ratio between the exon 15-16 primer set/exon 8-10 primer set was calculated to determine the relative expression level of the USP5 isoform 1. Reaction consisted of 10 μl of SYBR Green PCR master mix, 1.5 μl of 4 μM stock solution of each the reverse and forward primers, and 7 μg of cDNA in a final reaction volume of 20 μl. All PCR assays were performed in triplicate. Master mixes and probes were purchased from Applied Biosystems (Foster City, CA), and the primers used for studying USP5 expression were purchased from Invitrogen. The SYBR Green primer sets included USP5 exon 15 5′-GCAACGAAGACGAAGACTCC-3′, USP5 exon 16 5′-CATAGGGAATCCCATCTCCA-3′, USP5 exon 8 5′-GAGTCAGGTGTGCCACTCAA-3′, and USP5 exon 10 5′-CCGGTACTGGCTTGGAATAC-3′.
UV Cross-Linking and Immunoprecipitation
UV cross-linking and immunoprecipitation was modified from a previously described method [23]. Briefly, IRDye® 800 end-labeled oligonucleotide (5′-ACTCTTCTTCCTGCCTGTCTCTCTCCCGTGCTGATGGGGG-3′)(Integrated DNA Technologies, Inc, Coralville, IA) was incubated with 400 μg whole cell lysate at 30°C for 10 min in a 50 μl reaction mixture. Reaction mixtures were transferred to ice and cross-linked with a Sylvania G115T8 lamp at a distance of 4 cm for 2.5 min, followed by RNase A treatment at 37°C for 10 min. The UV cross-linked complexes were immunoprecipitated using PTB-specific monoclonal antibody DH7 (Invitrogen) or normal mouse IgGs (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) as previously described [24]. Bound proteins were recovered in 30 μl SDS sample buffer, separated on a 12% SDS-polyacrylamide gel, and visualized using the Odyssey imaging system (Li-Cor Biosciences, Lincoln, NE) [19].
Morpholino Oligonucleotide Treatment
Morpholino oligonucleotides were purchased from Gene Tools, LLC (Philomath, OR). The 15ss morpholino (5′-AGAAACCAAGGCTACCTTTGGGCTC-3′) was designed to hybridize to the 5′ splice site of exon 15 used to produce isoform 2, thus preventing the recognition of this splice site. A second morpholino targeted a predicted PTBP1 binding site within intron 15 (5′-GGGAGAGAGACAGGCAGGAAGAAGA-3′). Morpholino treatment was performed as previously described using scrape loading [25]. Briefly, cells were initially plated at 1 × 106 cells into a 6-cm dish and allowed to attach overnight. The following day, the medium was removed, the cells were washed once with phosphate-buffered saline, and then 1.5 ml medium containing 2.5 μM morpholino was added. The cells were detached from the dish using a cell scraper (Sarstedt cat. # 83.1830, Newton, NC), resuspended, and replated into a 6-cm dish. A second morpholino treatment was performed 72 hours later. Twenty-four hours after the second treatment, RNA was collected for analysis. For cell proliferation and wound-healing studies at the time of the second treatment, the cells were placed in 24-well plates.
Cell Proliferation and Migration Assay
Growth curves were generated using the standard culture method, with the inclusion of 1% fetal bovine serum. In brief, immediately following the second morpholino oligonucleotide treatment, 10,000 U251 cells or 6,500 LN229 cells were plated into individual wells of a 24-well plate (day 0). To determine cell counts, we used a hemacytometer taking the average cell number derived from four separate wells ± the standard error. Cell migration was monitored using a scratch assay as previously described [19]. Immediately following the second morpholino treatment, cells were plated into a 24-well plate at 250,000 cells per well. Twenty-four hours after plating, the cells were checked for confluency. At this point, a scratch was made using a 200-μl pipette tip (day 0). Two or three scratches were made per well. On this day, the medium was replaced to reduce the serum concentration from 10% to 1%. Cell migration was photographically monitored every 24 hours until the wound closed. To measure cell migration, we used the magic wand tool in Adobe Photoshop CS3 version 10 (San Jose, CA) with a tolerance of 3. Cell migration was reported as a measure of the percentage of the gap filled by taking the ratio of individual time points/day 0.
Bioinformatic and Statistical Analysis
Data analysis for exon arrays was done using GeneSpring GX software. The CEL files described above were loaded into GeneSpring GX using the ExonRMA16 summarization algorithm ExonRMA16 for core transcript and probesets with quantile normalization and baseline transformation to the median of all samples. We reported PTBP1 expression levels as the log normalized value for transcript cluster ID 3815165. For comparative analysis, study datasets were derived in a stepwise manner using the GeneSpring GX exon splicing analysis software tools. Filtering was performed first using the detection above background (DABG) option to eliminate low signal, high background, and cross-hybridization potential. Splicing analysis of variance (ANOVA) was next performed to calculate gene-level normalized intensities to ensure detection of significant probeset differences between groups. Finally, a Splicing Index (SI) parameter was applied to identify transcripts with individual probesets that differed significantly between groups from the gene probeset-normalized intensities. Splicing visualizations were created using the mean values normalized intensity +/− standard error derived for each probeset. Pearson analysis was performed on exported probeset data for USP5 transcript cluster ID 3402899 and PTBP1 transcript cluster ID 3815165 using Prizm 5v5.0c for Mac OS X (GraphPad Software, Inc., La Jolla, CA). For each sample, the USP5 isoform 1 level was represented by the ratio of the value for probeset ID 3402925 divided by the mean of the remaining probesets for the transcript cluster. The PTBP1 level was represented by the log of the mean value for all probesets comprising the transcript cluster. The PTBP1-binding sites were derived by uploading the custom track found at http://fairbrother.biomed.brown.edu/data/SelexMap/PTBNoLimitScaled.wig.gz [26] on to the May 2004 (NCBI35/hg17) assembly in the UCSC Genome Bioinformatics Browser Web site (http://genome.ucsc.edu/). Data for the USP5 gene were downloaded as PostScript files to generate figures.
RESULTS
Identification of PTBP1-regulated RNA Splicing
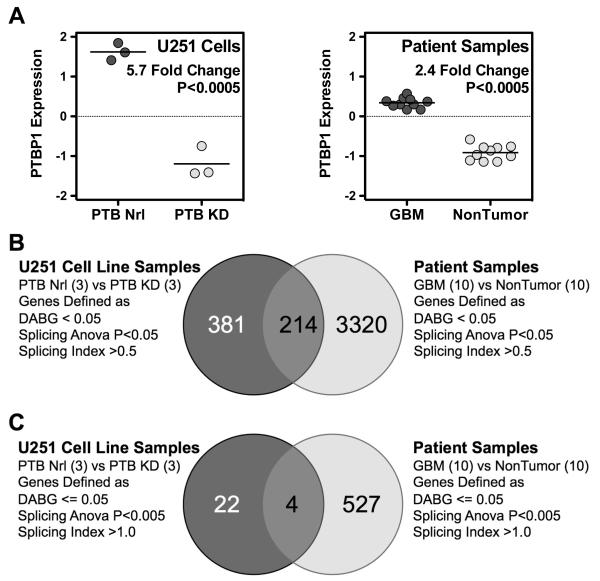
It is clear that alternative RNA splicing is a pervasive mechanism available to generate unique cellular transcripts. Regulation of alternative splicing thus plays a critical role in normal cellular function. We previously demonstrated that the splicing regulator PTBP1 is aberrantly upregulated in GBMs and that these tumors express multiple alternatively spliced RNA transcripts [8,18]. The splicing regulator PTBP1 is a 57-kDa protein belonging to the hnRNP protein family, whose members usually function as repressors of exon recognition during splicing [27-29]. The protein contains four RNA-recognitions motifs, an N-terminal nuclear localization signal, and a nuclear export signal [30,31]. In addition to playing a part in splicing repression, PTBP1 plays roles in exon inclusion, mRNA stability, RNA transport, viral translation and replication, 3′ end processing, and cap-independent translation through internal ribosomal entry site sequences (Reviewed in [31]). To define PTBP1-mediated, GBM-specific splicing events, we performed exon array analysis on U251 GBM cells with and without PTBP1 knockdown [19]. Surprisingly, this analysis revealed only a single PTBP1-specific splicing change when strict criteria were applied. Our results differed from the findings of array-based studies in N2A and HeLa cells, in which multiple RNA targets of PTBP1 action have been described [32-34]. These differences may have resulted from cell line-specific gene transcription, cell-specific splicing regulation, the level of stringency we applied in our analysis, or a combination of these factors. Therefore, we decided to reexamine our criteria for defining glioma-specific splicing events regulated by PTBP1. To do this, we performed comparative studies relying on array datasets from previous studies and bioinformatic tools available in the GeneSpring GX software suite. Patient array datasets were selected to maximize differential PTBP1 expression. We selected 10 nontumor samples and 10 GBM samples with an average 2.4-fold difference in PTBP1 expression (Figure 1A). This compares to a 2.9-fold difference for a 5 nontumor versus 5 GBM sample comparison (data not shown) and a 5.7-fold expression difference seen following PTBP1 knockdown in U251 cells (Figure 1A). We then used the RNA splicing tools available in the GeneSpring GX software package to generate lists of candidate PTBP1 targets. To remove genes with undetectable or low expression, we performed a default DABG filter analysis. Genes were kept on the basis of core probe selection region (PSR) data returning transcripts with at least 50% of the transcript probes present above background and a P value of <0.05. This subset was then used to identify genes with splicing ANOVA P values <0.05 and finally derive transcripts within this group with an absolute Splicing Index (SI) greater than 0.5. The splicing ANOVA and Splicing Index tools function to identify differential hybridization levels between probesets following gene-level normalization. The splicing ANOVA determines statistically significant probeset differences while the Splicing Index provides a log-based fold change difference (SI = 1 represents a 2-fold normalized hybridization difference). The analysis defined 595 transcripts in the U251 cell sample set and 3534 transcripts in the patient sample set. As has been observed in other studies [32-34], the results imply that there are hundreds of splicing events that depend on the presence of PTBP1. However, the data further suggest that only a portion of GBM-specific splicing is regulated by PTBP1. The overlap between the two groups was 214, representing 40% of the U251 and 6% of the patient transcripts, respectively (Figure 1B).
Figure 1.
Identification of PTBP1-mediated RNA splicing targets. (A) PTBP1 expression levels derived from Affymetrix exon array data. Left graph displays values for U251 GBM cells treated with control (PTB Nrl) or knockdown (PTB KD) morpholinos. Right graph shows the PTBP1 expression values for the 10 GBM samples and 10 nontumor samples used in this study. (B) and (C) Venn diagrams depicting the statistical derivation of genes with PTBP1-mediated changes in splicing. Datasets defined in (A) were used to derive PTBP1 target genes as defined by the indicated GeneSpring GX statistical parameters.
In performing this comparative analysis, we expected the patient array data to return a greater number of differential splicing events because of the cell heterogeneity that exists in tissue. Our hope was that the comparison with the target set derived from U251 cells would help limit these datasets to the most relevant genes specific to GBM cells. First, we were surprised to see that 381 transcripts identified as PTBP1-regulated in the U251 cell line were absent from the patient dataset. A direct examination of these genes in the patient dataset revealed that they were absent because they failed at least one of the default cutoff parameters used (103 failed DABG, 163 failed splicing ANOVA, and 115 failed the splicing index). A greater concern, however, was that on closer examination, we found that only 121 of the 214 transcripts identified as common events actually had Splicing Index values were both positive or negative (in our analysis a positive Splicing Index was associated with a reduced hybridization signal in the presence of high PTBP1 expression). Using a more stringent splicing ANOVA cutoff (P <0.005) reduced the identification of events with differential hybridization signals from 43% to 29%; alternatively, increasing the Splicing Index (SI>1) reduced the number of events from 43% to 20% (data not shown). When both conditions were applied, the composition of the groups greatly changed. The number of U251 cell PTBP1-targeted transcripts decreased by 65% from 595 to 26, while in the patient group, the transcript number decreased by 83% (from 3534 to 531; Figure 1C). Somewhat unexpectedly, the frequency of genes present in the intersection between the two groups decreased rather than increased. In the U251 cell line, only 15% (4 of 26) of the genes predicted to be regulated by PTBP1 were identified in the patient group (Figure 1C). When we examined the reason for this reduction, 10 of the 26 samples had SI values in the opposite direction of that observed for the transcripts in the patient group. Another three samples were eliminated because the splicing ANOVA P value was greater than 0.05. Somewhat surprising was the fact that 7 of the remaining 13 transcripts were found to involve different exons when we performed splicing visualizations comparing the two groups (data not shown). Thus, while the use of GeneSpring GX clearly helped to identify several hundred transcripts as potentially regulated by PTBP1 in each group, a comparative analysis using only the primary statistical tools supplied by the software suggested that biologically relevant events may be rare and therefore of greater significance.
Comparative Analysis Aids in the Identification of Regulated Splicing Events
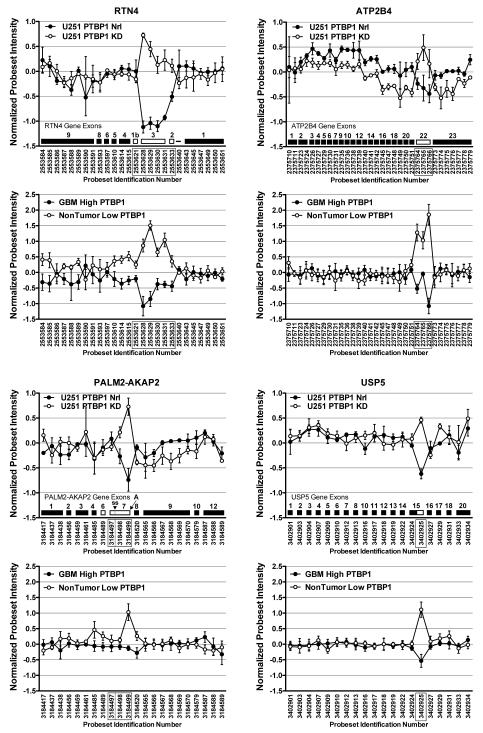
A more detailed examination of the hybridization profiles for the four gene transcripts identified by the comparative analysis is provided in Figure 2. This analysis confirms that for each gene, the “array-defined” splicing events involve similar PSRs in the U251 cells lines and the patient samples. Consistent with the role of PTBP1 as a splicing repressor, exon skipping was seen for three genes (RTN4, ATP2B4, and PALM2-AKAP2), and occlusion of a 5′ splice site was seen for the remaining gene (USP5). Two of the genes, RTN4 and ATP2B4, have been shown to be regulatory targets of PTBP1 [19,33-35]. We previously demonstrated that PTBP1-mediated RTN4 exon 3 skipping is associated with enhanced proliferation of GBM cells in culture [19]. The ATP2B4 gene was included among array-detected genes regulated by PTBP1 in the mouse N2A neuroblastoma cell line [33,34] as well as GBM-specific splicing [36]. Our array and RT-PCR data (not shown) validate ATP2B4 exon 22 splicing regulation by PTBP1 in human GBM (Figure 2).
Figure 2.
Identification of individual exon splicing events regulated by PTBP1. Probeset hybridization plots were generated for the four genes, RTN4, ATP2B4, PALM2-AKAP2, and USP5, identified in Figure 1C. For each gene, the upper U251 dataset graph provides individual Affymetrix probeset identification numbers with exons schematically illustrated. Alternative splicing events are denoted by unfilled exons. The alternative event examined in the PALM2-AKAP2 gene involves the choice between a 5′ splice site (ss) and a polyadenylation site (A) in exon 7. The probesets denoting the PTBP1-regulated events are boxed. The lower graphs provide data derived from the patient sample arrays.
Notably absent from our gene list was PKM2. The exclusion of exon 9 of the PKM2 gene has been demonstrated to be PTBP1-dependent in HeLa and 293 cells [21,28]. Furthermore, regulated mutual exclusion of exons 9 and 10 appears to highly correlate with the PTBP1 expression levels in gliomas [21]. Indeed, we also observed this splicing regulation in our patient array datasets (see Supplemental Figure 1). Our GBM samples with high PTBP1 showed exon 9 skipping with simultaneous exon 10 inclusion. In nontumor samples expression low PTBP1 the opposite splicing pattern was observed, exon 9 inclusion and exon 10 skipping. Surprisingly, there was no effect of PTBP1 depletion on PKM2 splicing in the U251 GBM cell lines. Exons 9 and 10 had similar inclusion levels independent of PTBP1 expression. This finding was confirmed by RT-PCR analysis that found predominant exclusion of exon 9 in U251 and LN229 glioblastoma cells even after PTBP1 knockdown (see Supplemental Figure 1). However, it is important to note that the only direct examination of PTBP1 knockdown in GBM cells (A172 cell line) revealed exon 9 inclusion increased from 6% to only 18% using semi-quantitative RT-PCR [20]. Thus, the direct role of PTBP-1 in GBM-specific PKM2 splicing regulation remains unclear.
PTBP1 regulation of PALM2-AKAP2 and USP5 splicing has not been previously described. For the PALM2-AKAP2 locus the predicted splicing is complicated by the fact that three independent transcripts derive from this locus, PALM2, AKAP2, and the PALM2-AKAP2 readthrough. RNA processing involving exon 7 plays a key role in the production of these three transcripts. The PALM2 transcript is produced when polyadenylation occurs at exon 7, detected by hybridization to Affymetrix PSRs 3184498 and 3184499. An AKAP2 mRNA is produced with the alternative use of a 5′-splicing site in exon 7, detected by hybridization only to Affymetrix PSR 3184497. Finally, the complete skipping of exon 7 results in the production of the PALM2-AKAP2 transcript, detected by lack of hybridization to all three Affymetrix PSRs. The array hybridization results are most consistent with PTBP1 regulating the choice between RNA splicing (high PTBP1 expression) and polyadenylation (low PTBP1 expression). Thus in GBM the production of AKAP2 mRNA would be favored. Neither event could be confirmed by RT-PCR (data not shown). It is unclear if this result can be attributed to array artifact (e.g., PSR cross-hybridization) or an inability to detect the low level expression of PALM2-AKAP2. In contrast to the other genes, USP5 demonstrated a consistent PTBP1-dependent exon hybridization profile between the two datasets despite the fact that it had the lowest average splicing ANOVA and Splicing Index scores of the four common genes. The PTBP1-dependent alternative 5′ splice site usage detected by PSR 3402925 was confirmed by RT-PCR analysis (Figure 3; data not shown). Furthermore, this splicing was recently reported in HeLa cells lacking both PTBP1 and PTBP2 [32].
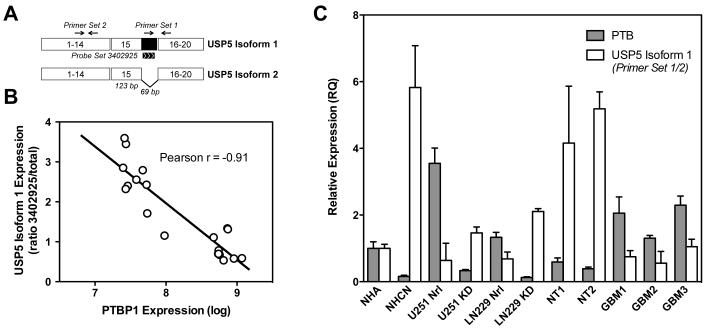
Figure 3.
Splicing of USP5 correlates with PTBP1 expression levels. (A) Schematic representation of the USP5 mRNA isoforms. Alternative use of a 5′ splice site within exon 15 removes 69 bases of coding sequence to generate isoform 2. The location of PCR primer sets and Affymetrix probeset 3402325 are indicated. (B) Plot of dataset values for USP5 splicing versus PTBP1 expression levels. (C) Validation of USP5 splicing was performed by quantitative RT-PCR as described in Materials and Methods. Samples included normal human astrocytes (NHA), normal human cortical neurons (NHCN), U251 and LN229 GBM cell lines treated with control morpholino (Nrl) or PTBP1 knockdown morpholino (KD), two nontumor brain samples (NT) and three GBM samples (GBM).
Alternative Splicing of USP5 Is Regulated by PTBP1
Because USP5 was a novel PTBP1 target, we chose to examine this splicing event in greater detail. Also known as isopeptidase T, USP5 encodes a highly conserved deubiquitinating enzyme [37]. In humans, alternative splicing is known to generate two mRNAs that differ by the inclusion (isoform 1) or exclusion (isoform 2) of the 69 nucleotides ending exon 15 (Figure 3A) [38]. When USP5 isoform 1 expression was plotted against PTBP1 expression for the 20 individual patient samples used in our array analysis, we obtained a significant correlation (Figure 3B). Expression of USP5 isoform 1 occurred with reduced PTBP1 levels. This result suggests that PTBP1 expression inhibits recognition of the isoform 1-specific 5′ splice site. To validate the array findings, we performed quantitative assessment of alternative splicing using quantitative RT-PCR. The relative expression of USP5 isoform 1 was determined as the ratio of mRNAs including the 69-nucleotide region over total USP5 mRNA (measured as exons 8-10). The quantitative RT-PCR analysis confirmed the exon array data; USP5 isoform 1 expression was upregulated in U251 cells with PTBP1 knockdown compared to control cells and in nontumor patient samples compared to GBM samples (Figure 3C). We also confirmed PTBP1-dependent regulation in a second GBM cell line, LN229, where knockdown resulted in a 3-fold upregulation in USP5 isoform 1 expression. In normal neural cells, USP5 splicing also paralleled PTBP1 expression levels. In cultured normal human cortical neurons, USP5 isoform 1 expression was significantly elevated compared to expression in normal human astrocytes (Figure 3C). The data support a role for PTBP1 in the regulation of USP5 splicing. High levels of PTBP1 are associated with the expression of USP5 isoform 2, while reduced PTBP1 is required for production of USP5 isoform 1.
Identification of Sequences that Regulate USP5 Splicing
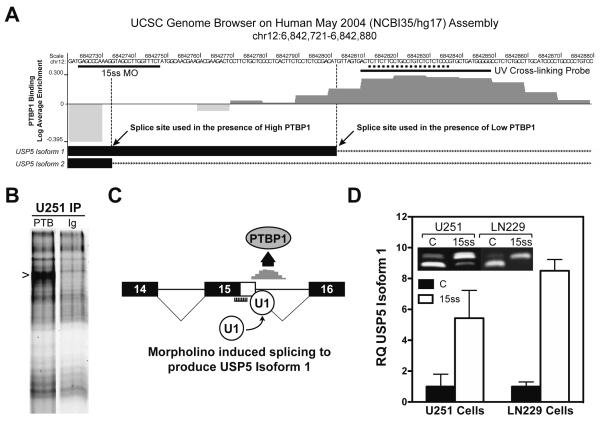
Our data clearly define a role for PTBP1 in USP5 splicing but do not address whether regulation is direct or indirect. To help clarify this regulatory role, we performed an in silico analysis to define PTBP1-binding sites. As a first step, we used the Splicing Rainbow program available at the EBI ASD Project Alternative Splicing Workbench to localize predicted PTBP1-binding sites for all four candidate genes [39]. Not surprisingly, the regions encompassing the alternative exons contained predicted PTBP1-binding consensus sequences (Supplemental Figure 2). Subsequently, we examined databases from two studies using genome-wide approaches to define PTBP1-binding sites in HeLa cells [26,40]. One study used in vivo CLIP-seq analysis to define RNA sequences bound by PTBP1 [40]. Although this study showed binding clusters in nearly 50% of annotated genes, we only identified PTBP1-binding sites for the PALM2-AKAP2 and RTN4 genes (Supplemental Figure 2). The frequency and location of PTBP1-binding sites were consistent with regulatory predictions for RTN4 splicing but not for PALM2-AKAP2 [29,32,40]. A limitation of CLIP-seq is that the identification of binding sites depends on transcript level. Among the four genes, only RTN4 is expressed at appreciable levels in HeLa cells (GDS1779 [41]). To overcome this issue, Reid et al. developed an in vitro SELEX method using a series of tiled 30-mer oligonucleotides encompassing ~4000 exons [26]. Among the four candidate genes only USP5 exon 15 was included among the exons identified by our analysis (Supplemental Figure 2). Figure 4A provides the experimentally derived PTBP1-binding frequency in HeLa nuclear extract for the region encompassing the alternatively used 5′-splice sites of USP5 [26]. Specific PTBP1-binding is seen at the 5′-splice site region used to generate the USP5 isoform 1 and not within the splice site region required for the production of isoform 2. We confirmed similar PTBP1-binding occurs in U251 cells by performing UV cross-linking to the target sequence (underlined) followed by specific immunoprecipitation of the complexes formed (Figure 4B). The binding pattern is consistent with the occlusion model of splicing regulation, in which PTBP1 binding prevents splice site recognition [29,40]. The findings also provide an opportunity to define the specific contribution of USP5 splicing to gliomagenesis by redirecting splice site selection independent of PTBP1. We previously demonstrated that antisense morpholino oligonucleotides can be used to block PTBP1 interactions and change RNA splicing [25]. A single morpholino oligonucleotide targeted to the peak PTBP1-binding region proved ineffective in changing splice site choice (Figure 4A; data not shown). It is unclear whether targeting only a portion of the region was unable to completely block PTBP1-binding or if the proximity of morpholino oligonucleotide-binding prevented normal splice site recognition. To circumvent the problem, we chose to target the USP5 isoform 2 splice site assuming that preventing U1 snRNP association would force recognition of the less-favored downstream splice site to exclude PTBP1 (Figure 4C). Treatment of U251 and LN229 GBM cells with 15 ss morpholino oligonucleotide caused a dramatic upregulation of USP5 isoform 1 expression as detected by either standard or quantitative RT-PCR (Figure 4D). In the LN229 cell line, there was a greater than 8-fold increase in USP5 isoform 1, clearly indicating that morpholino oligonucleotide treatment was able to alter splice site choice in the presence of PTBP1.
Figure 4.
Identification of PTBP1-binding sites and modulation of USP5 RNA splicing. (A) Schematic representation of SELEX-derived PTBP1-binding sites using HeLa cell nuclear extract within the USP5 alternatively regulated region [38]. The graph displays sequences found enriched (dark grey) or underrepresented (light grey) when examined for PTBP1 binding. The peak PTBP1-binding region maps from 6842820 to 6842829. PTBP1 binding is distinctly absent from the USP5 isoform 2 5′ splice site region. The UCSC Browser download for the genomic region indicated was modified to include only relevant information. The hybridization sites for the morpholino oligonucleotides and the sequence used to generate the UV cross-linking probe used in (B) are indicated. (B) UV cross-linking of U251 cell lysate with USP5 intron 15 IR800-endlabeled probe. Cross-linked complexes were immunoprecipitated (IP) with PTBP1 antibody (PTB) or antimouse immunoglobulin antibody (Ig), separated by SDS polyacrylamide gel electrophoresis and visualized by IR scanning. The PTBP1-specific band is noted. (C) Schematic illustration of the predicted outcome derived from the hybridization of 15 ss morpholino to USP5 precursor RNA. (D) Quantitative and semiquantitative (inset) RT-PCR analysis of U251 and LN229 GBM cells treated with control (C) or 15 ss morpholino.
USP5 Isoform 1 Production Is Associated with Reduced GBM Cell Growth and Motility
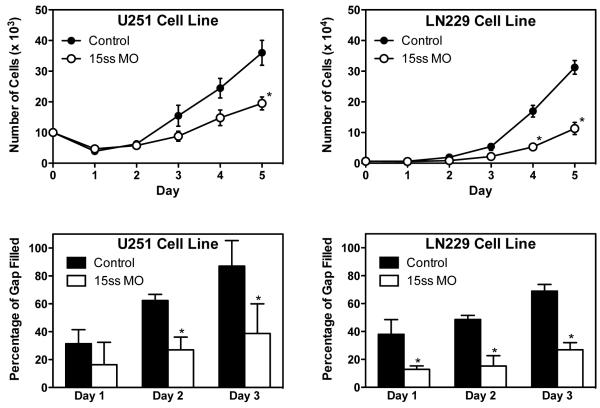
The ability to target USP5 splicing independent of PTBP1 expression level provides a direct mechanism by which to assess the functional role of isoform 2 in GBM cells. We previously demonstrated that PTBP1 knockdown inhibits the growth and migration of U251 and LN229 GBM cells [19]. The ability of PTBP1 to inhibit cell proliferation was found to be in part due to the RNA splicing regulation of RTN4. Ectopic expression of the RTN4 isoform containing exon 3 was found to reduce U251 cell growth; however, it had no effect on cell migration [19]. Treatment of U251 and LN229 GBM cells with 15 ss morpholino oligonucleotide to increase USP5 isoform 1 production was found to inhibit both cell proliferation and migration (Figure 5). The inhibitory effect was greater in LN229 cells than in U251 cells, consistent with the ability of 15 ss morpholino oligonucleotide to induce a greater expression of USP5 isoform 2. Cell proliferation at 5 days was reduced by 46% in U251 cells and 64% in LN229 cells by treatment with 15 ss morpholino oligonucleotide. A similar trend was observed for cell migration. We also observed that LN229 cells were slower to fill the scratch-induced gap than U251 cells.
Figure 5.
Modulation of USP5 splicing to generate isoform 1 is associated with growth and migration reductions in GBM cells. U251 and LN229 GBM cells were treated with control or 15 ss morpholino as described in Materials and Methods. Cells were monitored for growth and migration as indicated in individual graphs. *P<0.05
DISCUSSION
There is a growing consensus that misregulation of RNA splicing plays an important role in tumorigenesis. We have studied gliomagenesis to define roles for specific mRNA isoforms in tumor growth and invasion [8,19,42]. An examination of splicing in GBM revealed the differential expression of several splicing regulatory factors and the production of tumor-specific mRNA isoforms [8]. Sorting out the contributions of specific factors and isoforms to tumorigenesis will likely prove a great challenge. In this study, we focused on identifying the GBM-specific splicing events that appear to be physiologically relevant targets of PTBP1 regulation. Our results are notable for the low number of gene targets uncovered, three (RTN4, ATP2B4, and USP5), and the absence of the previously reported PTBP1 target PKM2 [20,21]. While GBM-specific splicing of PKM2 was observed in patient tumors, regulated splicing was not uniquely dependent on PTBP1 levels. In U251 and LN229 glioblastoma cells with PTBP1 knockdown, PKM2 exon 9 was still skipped, and in individual patient samples we observed that PKM2 splicing did not correlate with PTBP1 level (Supplemental Figure 1; data not shown). It is possible that splicing regulation may differ in neural cells and/or depends on changes in regulators other than those we studied. Within our complete 36-patient Affymetrix exon array dataset, expression of RBM9 (r = −0.66) and SF3A2 (r = 0.61) showed the strongest correlations with exon 9 splicing. Whether these splicing factors or others play a role in GBM-specific splicing of PKM2 remains to be addressed.
A major function of PTBP1 is to function as a repressor of exon inclusion during RNA splicing [29,32,40]. The tissues of the adult brain is one of the areas where PTBP1 is expressed at low levels [43]; thus, elevated PTBP1 expression coupled with the aberrant expression of other regulators in GBM should dramatically alter the isoform composition of expressed RNAs. While the aberrant expression of splicing regulators has been proposed as one mechanism that fuels tumorigenesis [6], few studies detail how aberrant mRNA isoform expression drives this process. Thus, questions remain regarding the contributions of individual splicing factors and splicing events to tumor cell proliferation, and what role functional redundancy may play for many splicing regulators with known homologues. PTBP2 is a functionally related homologue that is upregulated in many cell types in the absence of PTBP1 [33]. Indeed, we have previously demonstrated that knockdown of PTBP1 in U251 cells is accompanied by upregulation of PTBP2 [19]. However, performing a similar comparative analysis using U251 cells with simultaneous PTBP1 and PTBP2 knockdown yielded only a single additional commonly regulated splicing target, TPM1 (data not shown). This would imply that identification of important PTBP1-regulated mRNA targets was not obscured by direct functional redundancy due to PTBP2, but does not rule out redundancy of other regulators (e.g. related hnRNPs). One of the best examples demonstrating the role of splicing in tumorigenesis is the SFRS1-mediated transformation of NIH 3T3 cells [15]. Although an unbiased genome-wide examination of RNA splicing was not performed, clear differences in mRNA isoform expression were reported. Among several regulatory events identified, cell transformation was recapitulated by a single RNA splicing event involving the specific expression of S6K1 isoform 2. These findings somewhat parallel our results with PTBP1 in GBM, where much of the tumorigenic phenotype is lost when either RTN4 [19] or USP5 splicing is corrected. Therefore, at least in these two examples, it may be appropriate to speculate that tumorigenesis is driven by a few specifically targeted events rather than a broad spectrum of splicing changes.
In this study, we showed for the first time that differential splicing of USP5 plays a role in GBM cell growth and motility. Although USP5 is thought to function primarily on unanchored branched polyubiquitin chains to generate ubiquitin monomers [37], the specific functional role of individual USP5 isoforms generated by alternative splicing remains unclear. The longer USP5 isoform 1 generates a protein with a substitution of Ala629 with 23 amino acids (Gly629 to Ser652). The insertion occurs between two ubiquitin binding domains (UBP and UBA1) which have been proposed to affect the binding of the fourth ubiquitin in the chain [44]. When the in vitro activity of the two isoforms is compared using recombinant proteins, isoform 1 has higher ubiquitin-AMC cleavage activity [45]. This would suggest that the elevated PTBP1 expression in GBM is associated with a decrease in USP5 activity. Two recent studies have demonstrated that the loss of USP5 stabilizes p53 [46,47]. This finding seems inconsistent with our data demonstrating that forced expression of the more active USP5 isoform 1 is associated with reduced cell growth (Figure 5). Unfortunately, it is unclear if the use of ubiquitin-AMC provides a true measure of enzymatic activity against branched ubiquitin substrate. Also, given our observation that USP5 alters GBM cell migration, it seems more likely that targets other than p53 are involved. Our results demonstrate a clear functional consequence associated with USP5 alternative splicing and suggest that its role in gliomagenesis is more important than previously recognized. Uncovering the specific mechanism and targets of USP5 individual isoform action may provide insight into the development of novel therapeutic approaches in the treatment of GBM.
Supplementary Material
Supplemental Figure 1. The regulation of PKM2 splicing is not dependent on PTBP1 in U251 GBM cells. Probeset hybridization plots were generated for the PKM2 gene using the two array datasets described in Figure 1. (A) A comparison of 10 GBM PTBP1 high samples and 10 nontumor PTBP1 low samples. The hybridization plot shows skipping of PKM2 exon 9 in GBM High PTBP1 samples and inclusion of exon 10. This pattern differs from that observed in the NonTumor Low PTBP1 samples where exon 9 is included and exon 10 is skipped. For both exons the hybridization differences between the two sample groups were significant (*P = 0.001). The table to the right provides a list of all splicing factors with a greater than 2-fold absolute expression difference between the PTBP1 high and low samples. (B) Comparison of the U251 cell line samples with and without PTBP1 knockdown. The hybridization plot shows skipping of PKM2 exon 9 and inclusion of exon 10 in both datasets despite a >5-fold reduction in PTBP1 expression. It is noteworthy that of only HSPA5 showed a significant expression difference between the two groups, which given its magnitude of change was unlikely to be relevant. (C) RT-PCR examination of PKM2 splicing in glioblastoma cell lines. RNA isolated from U251 and LN229 glioblastoma cell lines with and without PTBP1 knockdown were directly assessed for PKM2 splicing as previously described [21]. Because the sizes of PKM2 exons 9 and 10 are equivalent the ratio of splicing products is determined by digestion with PstI digestion, which cuts within exon 10. Similar to the result observed in (B), RT-PCR found predominant exon 9 skipping in U251 cells, and in a second cell line LN229, with no effect of PTBP1 knockdown to enhance inclusion.
Supplemental Figure 2. Mapping of PTBP1-binding sites for the four genes, ATP2B4, PALM2-AKAP2, RTN4, and USP5, identified in Figure 1C. Diagrams are modified from UCSC Browser downloads showing the track information generated for PTBP1 blat derived [36], Clip-Seq [37], and SELEX [38] in silico approaches. Note that SELEX and Clip-Seq data was not available for all genetic regions shown.
ACKNOWLEDGMENTS
We thank Markeda Wade for assistance with the preparation of the manuscript.
This work was supported by NIH NCI CA67946 (to G.J.C.); The John K. Funk Endowment (to G.J.C.), Rosalie B. Hite Foundation (to H.C.C.) and the Kleberg Foundation (to R.K.), with additional support provided by the NIH through MD Anderson’s Cancer Center Support Grant CA016672.
Abbreviations
- PTBP1
polypyrimidine tract-binding protein 1
- ATP2B4
ATPase, Ca++ transporting, plasma membrane 4
- RTN4
reticulon 4
- PKM2
pyruvate kinase, muscle
- USP5
ubiquitin specific peptidase 5
- PALM2-AKAP2
paralemmin 2-A kinase (PRKA) anchor protein 2 readthrough
- GBM
glioblastoma multiforme
- SI
splicing index
- DABG
detection above background
- PSR
probe selection region
- SELEX
systematic evolution of ligands by exponential enrichment
REFERENCES
- 1.CBTRUS . Volume 2009. Central Brain Tumor Registry of the United States; 2009. http://www.cbtrus.org/ [Google Scholar]
- 2.Cancer Genome Atlas Research N Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin. 2010;60:166–193. doi: 10.3322/caac.20069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 5.Wang ET, Sandberg R, Luo S, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Srebrow A, Kornblihtt AR. The connection between splicing and cancer. Journal of Cell Science. 2006;119:2635–2641. doi: 10.1242/jcs.03053. [DOI] [PubMed] [Google Scholar]
- 7.Kirschbaum-Slager N, Lopes GM, Galante PA, Riggins GJ, de Souza SJ. Splicing factors are differentially expressed in tumors. Genetics and Molecular Research. 2004;3:512–520. [PubMed] [Google Scholar]
- 8.Cheung H, Baggerly K, Tsavachidis S, et al. Global analysis of aberrant pre-mRNA splicing in glioblastoma using exon expression arrays. BMC Genomics. 2008;9:216. doi: 10.1186/1471-2164-9-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao Y, Fotovati A, Lee C, et al. Inhibition of Y-box binding protein-1 slows the growth of glioblastoma multiforme and sensitizes to temozolomide independent O6-methylguanine-DNA methyltransferase. Mol Cancer Ther. 2009;8:3276–3284. doi: 10.1158/1535-7163.MCT-09-0478. [DOI] [PubMed] [Google Scholar]
- 10.Grosso AR, Martins S, Carmo-Fonseca M. The emerging role of splicing factors in cancer. EMBO Rep. 2008;9:1087–1093. doi: 10.1038/embor.2008.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venables JP. Aberrant and alternative splicing in cancer. Cancer Res. 2004;64:7647–7654. doi: 10.1158/0008-5472.CAN-04-1910. [DOI] [PubMed] [Google Scholar]
- 12.Schwerk C, Schulze-Osthoff K. Regulation of apoptosis by alternative pre-mRNA splicing. Mol Cell. 2005;19:1–13. doi: 10.1016/j.molcel.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 13.He X, Arslan AD, Pool MD, et al. Knockdown of splicing factor SRp20 causes apoptosis in ovarian cancer cells and its expression is associated with malignancy of epithelial ovarian cancer. Oncogene. 2010 Sep 20; doi: 10.1038/onc.2010.426. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He X, Pool M, Darcy KM, et al. Knockdown of polypyrimidine tract-binding protein suppresses ovarian tumor cell growth and invasiveness in vitro. Oncogene. 2007;26:4961–4968. doi: 10.1038/sj.onc.1210307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karni R, de Stanchina E, Lowe SW, Sinha R, Mu D, Krainer AR. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nature Structural and Molecular Biology. 2007;14:185–193. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu R, Heaney J, Nadeau JH, Ali S, Matin A. Deficiency of splicing factor 1 suppresses the occurrence of testicular germ cell tumors. Cancer Res. 2010;70:7264–7272. doi: 10.1158/0008-5472.CAN-10-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang C, Norton JT, Ghosh S, et al. Polypyrimidine tract-binding protein (PTB) differentially affects malignancy in a cell line-dependent manner. Journal of Biological Chemistry. 2008;283:20277–20287. doi: 10.1074/jbc.M803682200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheung HC, Corley LJ, Fuller GN, McCutcheon IE, Cote GJ. Polypyrimidine tract binding protein and Notch1 are independently re-expressed in glioma. Mod Pathol. 2006;19:1034–1041. doi: 10.1038/modpathol.3800635. [DOI] [PubMed] [Google Scholar]
- 19.Cheung HC, Hai T, Zhu W, et al. Splicing factors PTBP1 and PTBP2 promote proliferation and migration of glioma cell lines. Brain. 2009:2277–2288. doi: 10.1093/brain/awp153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clower CV, Chatterjee D, Wang Z, Cantley LC, Vander Heiden MG, Krainer AR. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc Natl Acad Sci U S A. 2010;107:1894–1899. doi: 10.1073/pnas.0914845107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 23.Jin W, McCutcheon IE, Fuller GN, Huang ES, Cote GJ. Fibroblast growth factor receptor-1 alpha-exon exclusion and polypyrimidine tract-binding protein in glioblastoma multiforme tumors. Cancer Res. 2000;60:1221–1224. [PubMed] [Google Scholar]
- 24.Lou H, Helfman DM, Gagel RF, Berget SM. Polypyrimidine tract-binding protein positively regulates inclusion of an alternative 3′-terminal exon. Mol Cell Biol. 1999;19:78–85. doi: 10.1128/mcb.19.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bruno IG, Jin W, Cote GJ. Correction of aberrant FGFR1 alternative RNA splicing through targeting of intronic regulatory elements. Hum Mol Genet. 2004;13:2409–2420. doi: 10.1093/hmg/ddh272. [DOI] [PubMed] [Google Scholar]
- 26.Reid DC, Chang BL, Gunderson SI, Alpert L, Thompson WA, Fairbrother WG. Next-generation SELEX identifies sequence and structural determinants of splicing factor binding in human pre-mRNA sequence. RNA. 2009;15:2385–2397. doi: 10.1261/rna.1821809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.García-Blanco MA, Jamison SF, Sharp PA. Identification and purification of a 62,000-dalton protein that binds specifically to the polypyrimidine tract of introns. Genes & development. 1989;3:1874–1886. doi: 10.1101/gad.3.12a.1874. [DOI] [PubMed] [Google Scholar]
- 28.Spellman R, Llorian M, Smith CWJ. Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Molecular Cell. 2007;27:420–434. doi: 10.1016/j.molcel.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wagner EJ, Garcia-Blanco MA. Polypyrimidine tract binding protein antagonizes exon definition. Mol Cell Biol. 2001;21:3281–3288. doi: 10.1128/MCB.21.10.3281-3288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oh YL, Hahm B, Kim YK, et al. Determination of functional domains in polypyrimidine-tract binding protein. Biochemical Journal. 1998;331:169–175. doi: 10.1042/bj3310169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sawicka K, Bushell M, Spriggs KA, Willis AE. Polypyrimidine-tract binding protein: a multifunctional RNA-binding protein. Biochemical Society Transactions. 2008;36:641–647. doi: 10.1042/BST0360641. [DOI] [PubMed] [Google Scholar]
- 32.Llorian M, Schwartz S, Clark TA, et al. Position-dependent alternative splicing activity revealed by global profiling of alternative splicing events regulated by PTB. Nat Struct Mol Biol. 2010;17:1114–1123. doi: 10.1038/nsmb.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boutz PL, Stoilov P, Li Q, et al. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xing Y, Stoilov P, Kapur K, et al. MADS: a new and improved method for analysis of differential alternative splicing by exon-tiling microarrays. RNA. 2008;14:1470–1479. doi: 10.1261/rna.1070208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.French PJ, Peeters J, Horsman S, et al. Identification of differentially regulated splice variants and novel exons in glial brain tumors using exon expression arrays. Cancer Res. 2007;67:5635–5642. doi: 10.1158/0008-5472.CAN-06-2869. [DOI] [PubMed] [Google Scholar]
- 37.Wilkinson KD, Tashayev VL, O’Connor LB, Larsen CN, Kasperek E, Pickart CM. Metabolism of the polyubiquitin degradation signal: structure, mechanism, and role of isopeptidase T. Biochemistry. 1995;34:14535–14546. doi: 10.1021/bi00044a032. [DOI] [PubMed] [Google Scholar]
- 38.Ansari-Lari MA, Muzny DM, Lu J, et al. A gene-rich cluster between the CD4 and triosephosphate isomerase genes at human chromosome 12p13. Genome Res. 1996;6:314–326. doi: 10.1101/gr.6.4.314. [DOI] [PubMed] [Google Scholar]
- 39.Stamm S, Ben-Ari S, Rafalska I, et al. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 40.Xue Y, Zhou Y, Wu T, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mense SM, Sengupta A, Zhou M, et al. Gene expression profiling reveals the profound upregulation of hypoxia-responsive genes in primary human astrocytes. Physiol Genomics. 2006;25:435–449. doi: 10.1152/physiolgenomics.00315.2005. [DOI] [PubMed] [Google Scholar]
- 42.Cote GJ, Bruno IG, Jin W. Abnormal regulation of RNA splicing in gliomas. In: Zhang W, Fuller GN, editors. Genomic and Molecular Neuro-Oncology. Jones and Bartlett Publishers, Inc; Sudbury: 2003. pp. 165–183. [Google Scholar]
- 43.McCutcheon IE, Hentschel SJ, Fuller GN, Jin W, Cote GJ. Expression of the splicing regulator polypyrimidine tract-binding protein in normal and neoplastic brain. Neuro Oncol. 2004;6:9–14. doi: 10.1215/S1152851703000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reyes-Turcu FE, Shanks JR, Komander D, Wilkinson KD. Recognition of polyubiquitin isoforms by the multiple ubiquitin binding modules of isopeptidase T. J Biol Chem. 2008;283:19581–19592. doi: 10.1074/jbc.M800947200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sciences EL. Isopeptidase T [UBP5] long form, (human, recombinant) product data sheet. 2009;Volume 2010 [Google Scholar]
- 46.Kapuria V, Peterson LF, Fang D, Bornmann WG, Talpaz M, Donato NJ. Deubiquitinase inhibition by small-molecule WP1130 triggers aggresome formation and tumor cell apoptosis. Cancer Res. 2010;70:9265–9276. doi: 10.1158/0008-5472.CAN-10-1530. [DOI] [PubMed] [Google Scholar]
- 47.Dayal S, Sparks A, Jacob J, Allende-Vega N, Lane DP, Saville MK. Suppression of the deubiquitinating enzyme USP5 causes the accumulation of unanchored polyubiquitin and the activation of p53. J Biol Chem. 2009;284:5030–5041. doi: 10.1074/jbc.M805871200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. The regulation of PKM2 splicing is not dependent on PTBP1 in U251 GBM cells. Probeset hybridization plots were generated for the PKM2 gene using the two array datasets described in Figure 1. (A) A comparison of 10 GBM PTBP1 high samples and 10 nontumor PTBP1 low samples. The hybridization plot shows skipping of PKM2 exon 9 in GBM High PTBP1 samples and inclusion of exon 10. This pattern differs from that observed in the NonTumor Low PTBP1 samples where exon 9 is included and exon 10 is skipped. For both exons the hybridization differences between the two sample groups were significant (*P = 0.001). The table to the right provides a list of all splicing factors with a greater than 2-fold absolute expression difference between the PTBP1 high and low samples. (B) Comparison of the U251 cell line samples with and without PTBP1 knockdown. The hybridization plot shows skipping of PKM2 exon 9 and inclusion of exon 10 in both datasets despite a >5-fold reduction in PTBP1 expression. It is noteworthy that of only HSPA5 showed a significant expression difference between the two groups, which given its magnitude of change was unlikely to be relevant. (C) RT-PCR examination of PKM2 splicing in glioblastoma cell lines. RNA isolated from U251 and LN229 glioblastoma cell lines with and without PTBP1 knockdown were directly assessed for PKM2 splicing as previously described [21]. Because the sizes of PKM2 exons 9 and 10 are equivalent the ratio of splicing products is determined by digestion with PstI digestion, which cuts within exon 10. Similar to the result observed in (B), RT-PCR found predominant exon 9 skipping in U251 cells, and in a second cell line LN229, with no effect of PTBP1 knockdown to enhance inclusion.
Supplemental Figure 2. Mapping of PTBP1-binding sites for the four genes, ATP2B4, PALM2-AKAP2, RTN4, and USP5, identified in Figure 1C. Diagrams are modified from UCSC Browser downloads showing the track information generated for PTBP1 blat derived [36], Clip-Seq [37], and SELEX [38] in silico approaches. Note that SELEX and Clip-Seq data was not available for all genetic regions shown.