Abstract
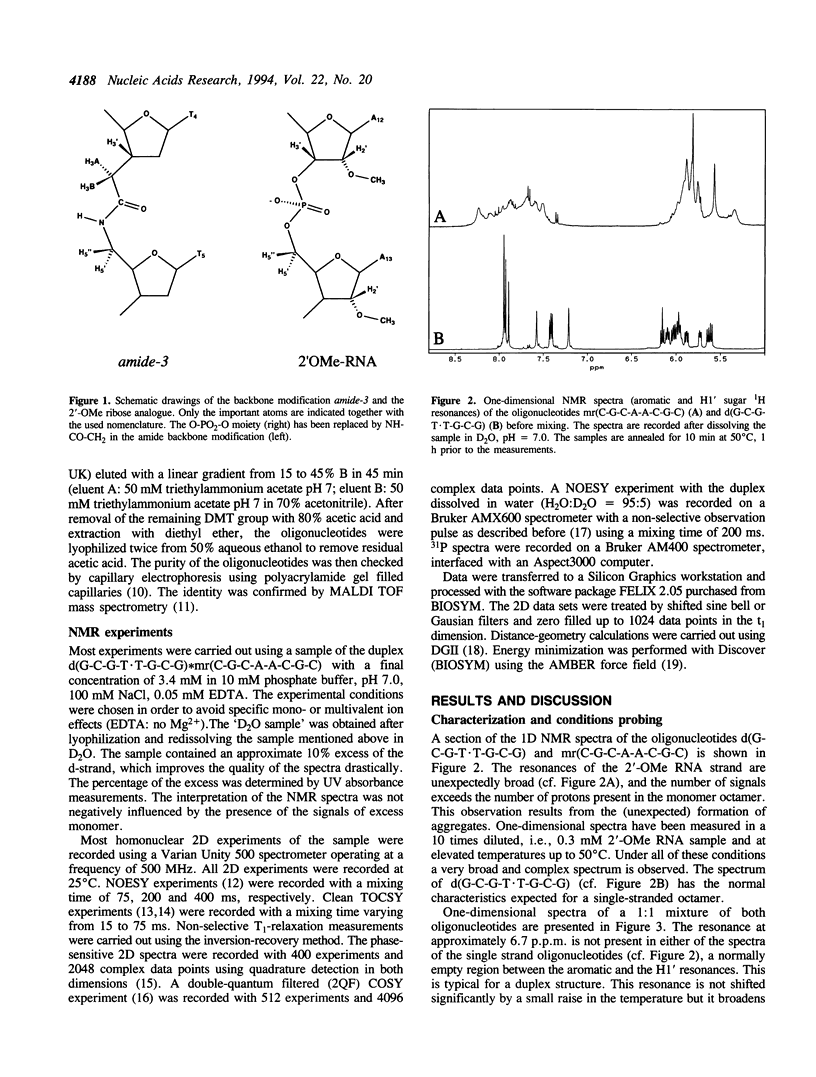
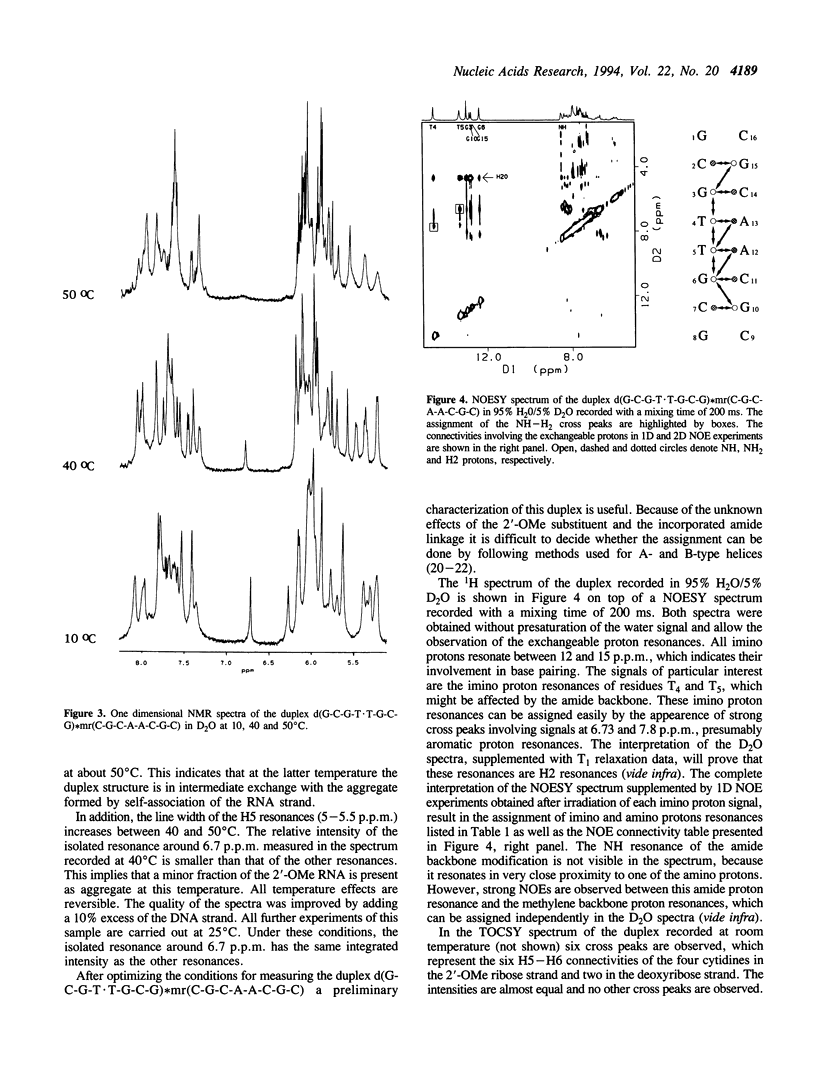
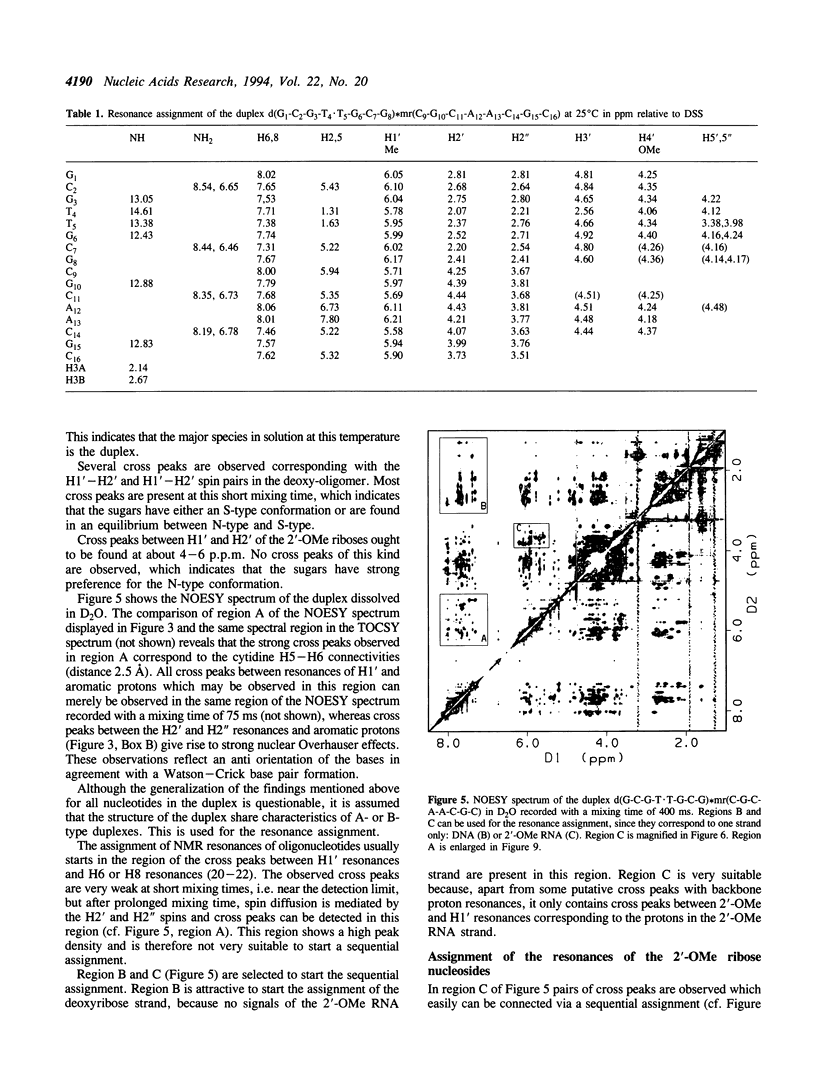
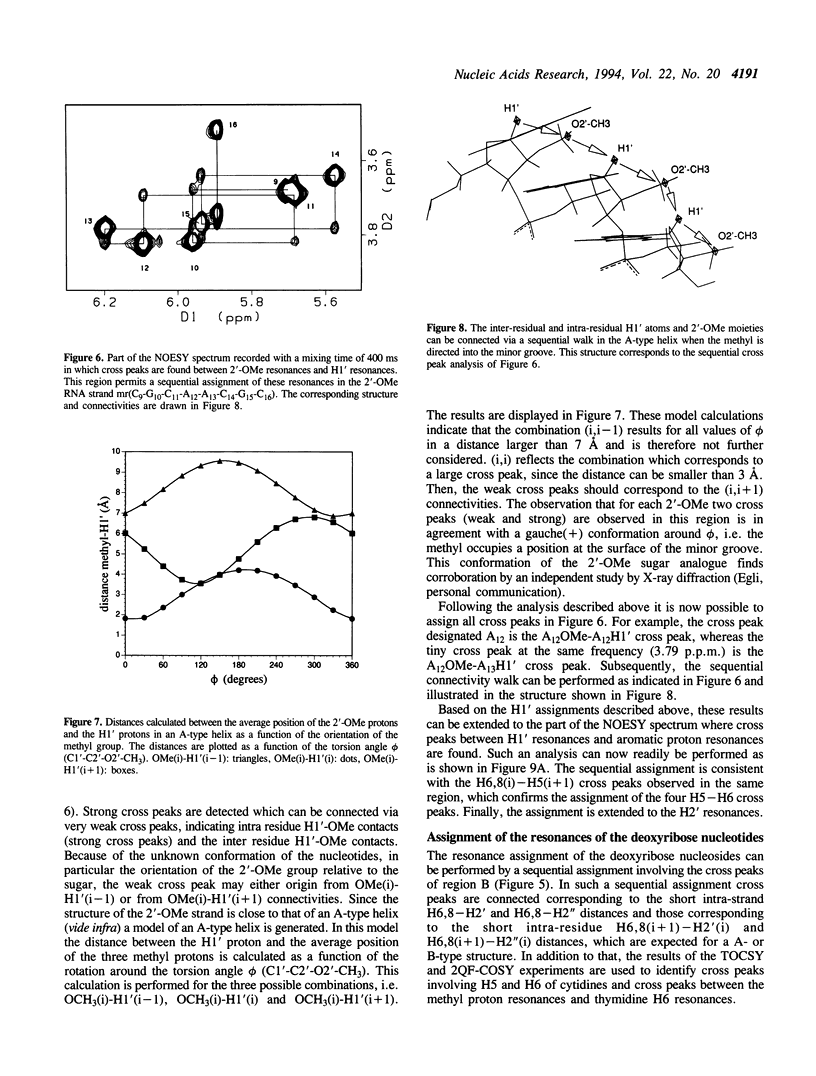
The backbone modification amide-3, in which -CH2-NH-CO-CH2- replaces -C5'H2-O5'-PO2-O3'-, is studied in the duplex d(G1-C2-G3-T4.T5-G6-C7-G8)*mr(C9-G10-C11-A12-A13-C14-G15+ ++-C16) where . indicates the backbone modification and mr indicates the 2'-OMe RNA strand. The majority of the exchangeable and non-exchangeable resonances have been assigned. The assignment procedure differs from standard methods. The methyl substituent of the 2'-OMe position of the RNA strand can be used as a tool in the interpretation. The duplex structure is a right-handed double helix. The sugar conformations of the 2'-OMe RNA strand are predominantly N-type and the 2'-OMe is positioned at the surface of the minor groove. In the complementary strand, only the sugar of residue T4 is found exclusively in N-type conformation. The incorporation of the amide modification does not effect very strongly the duplex structure. All bases are involved in Watson-Crick base pairs.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Gao X., Brown F. K., Jeffs P., Bischofberger N., Lin K. Y., Pipe A. J., Noble S. A. Probing structural factors stabilizing antisense oligonucleotide duplexes: NMR studies of a DNA.DNA duplex containing a formacetal linkage. Biochemistry. 1992 Jul 14;31(27):6228–6236. doi: 10.1021/bi00142a009. [DOI] [PubMed] [Google Scholar]
- Gao X., Jeffs P. W. Unusual conformation of a 3'-thioformacetal linkage in a DNA duplex. J Biomol NMR. 1994 Jan;4(1):17–34. doi: 10.1007/BF00178333. [DOI] [PubMed] [Google Scholar]
- Haasnoot C. A., Hilbers C. W. Effective water resonance suppression in 1D- and 2D-FT-1H-NMR spectroscopy of biopolymers in aqueous solution. Biopolymers. 1983 May;22(5):1259–1266. doi: 10.1002/bip.360220502. [DOI] [PubMed] [Google Scholar]
- Haasnoot C. A., Westerink H. P., van der Marel G. A., van Boom J. H. Conformational analysis of a hybrid DNA-RNA double helical oligonucleotide in aqueous solution: d(CG)r(CG)d(CG) studied by 1D- and 2D-1H NMR spectroscopy. J Biomol Struct Dyn. 1983 Oct;1(1):131–149. doi: 10.1080/07391102.1983.10507430. [DOI] [PubMed] [Google Scholar]
- Hare D. R., Wemmer D. E., Chou S. H., Drobny G., Reid B. R. Assignment of the non-exchangeable proton resonances of d(C-G-C-G-A-A-T-T-C-G-C-G) using two-dimensional nuclear magnetic resonance methods. J Mol Biol. 1983 Dec 15;171(3):319–336. doi: 10.1016/0022-2836(83)90096-7. [DOI] [PubMed] [Google Scholar]
- Havel T. F. An evaluation of computational strategies for use in the determination of protein structure from distance constraints obtained by nuclear magnetic resonance. Prog Biophys Mol Biol. 1991;56(1):43–78. doi: 10.1016/0079-6107(91)90007-f. [DOI] [PubMed] [Google Scholar]
- Pieles U., Zürcher W., Schär M., Moser H. E. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry: a powerful tool for the mass and sequence analysis of natural and modified oligonucleotides. Nucleic Acids Res. 1993 Jul 11;21(14):3191–3196. doi: 10.1093/nar/21.14.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. C., Kim S. G., Flynn P. F., Chou S. H., Orban J., Reid B. R. Errors in RNA NOESY distance measurements in chimeric and hybrid duplexes: differences in RNA and DNA proton relaxation. Biochemistry. 1992 Apr 28;31(16):3940–3946. doi: 10.1021/bi00131a008. [DOI] [PMC free article] [PubMed] [Google Scholar]


