Abstract
The region responsible for replication of Vibrio cholerae chromosome II (chrII) resembles those of plasmids that have repeated initiator binding sites (iterons) and an autorepressed initiator gene. ChrII has additional features: Its iterons require full methylation for initiator (RctB) binding, which makes them inactive for a part of the cell cycle when they are hemi-methylated. RctB also binds to a second kind of site, called 39-mers, in a methylation independent manner. This binding is inhibitory to chrII replication. The site that RctB uses for autorepression has not been identified. Here we show that a 29-mer sequence, similar to the 39-mers, serves as that site, as we find that it binds RctB in vitro and suffices to repress the rctB promoter in vivo. The site is not subject to methylation and is likely to be active throughout the cell cycle. The 29-mer, like the 39-mers, could inhibit RctB-dependent mini-chrII replication in Escherichia coli, possibly by coupling with iterons via RctB bridges, as was seen in vitro. The 29-mer thus appears to play a dual role in regulating chrII replication: one independent of the cell cycle, the other dependent upon iteron methylation, hence responsive to the cell cycle.
Keywords: Vibrio cholerae, Secondary chromosome, Transcriptional autorepression, Operator site, Replication control, Hetero-handcuffing
1. Introduction
About 10% of bacteria whose DNA has been sequenced are found to have divided their genome into multiple chromosomes (Harrison et al., 2010, Egan et al., 2005). How the chromosomes are maintained remains largely unknown, except in the case of Vibrio cholerae. The genome of this bacterium is divided into two chromosomes (chrI and chrII), which are maintained by separate programs, both for replication and segregation (Heidelberg et al., 2000, Egan & Waldor, 2003, Fogel & Waldor, 2006). There are also indications that the maintenance of the two chromosomes might be coordinated (Rasmussen et al., 2007, Val et al., 2008).
Here we are concerned with chrII replication control. Great strides in understanding chromosome replication in V. cholerae were possible primarily because of the nearly identical replication systems of chrI and the Escherichia coli chromosome, and the similarity of the chrII replication system to that of plasmids that have iterons (Egan & Waldor, 2003, Chattoraj, 2000). Iterons bind plasmid-encoded replication initiator proteins, and the binding is essential for initiation as well as controlling the initiation rate. Whereas the replication of chrI is regulated by the universal bacterial initiator DnaA (Duigou et al., 2006), replication of chrII is regulated by its own initiator RctB, which binds to the chrII iterons (Duigou et al., 2006, Pal et al., 2005). The chrII iterons are distinguished from plasmid iterons in having GATC sequences in them, whose adenine residue is methylated by Dam (Egan & Waldor, 2003). Methylation of both the strands of duplex DNA is required for efficient RctB binding, which is one reason why Dam is essential for V. cholerae survival (Demarre & Chattoraj, 2010, Julio et al., 2001). The methylation requirement also makes the iterons ineffective following replication, when they become hemimethylated. The iterons stay hemimethylated for about two-thirds of the cell cycle; prevention of initiator binding thus could be a major regulatory event to prevent premature reinitiation of replication.
It was realized early on that transcription of the chrII initiator gene rctB is repressed by the product of the same gene (autorepression), like the transcription of many plasmid initiator genes (Egan et al., 2006, Pal et al., 2005). The mechanism of autorepression remained obscure since unlike the situation in plasmids, no iterons could be recognized in the vicinity of the promoter of rctB. Recently, RctB was found to bind a second kind of specific site, called 39-mers (Venkova-Canova & Chattoraj, 2011). These sites are key to the negative control of chrII replication. Here we report that a 29-mer site, which has the conserved features of the 39-mers, is the one used by RctB for autorepression. The 29-mer additionally could contribute to the negative regulation of chrII replication, possibly via interactions with the origin iterons, as 39-mers do. The 29-mer thus controls replication indirectly by controlling transcription of the initiator gene and possibly directly by inhibiting origin function.
2. Methods
2.1 Bacterial strains and plasmids
V. cholerae and E. coli strains, and plasmids used in this study are listed in Table 1. For cloning, the fragments were obtained either by PCR amplification of N16961 (CVC205) DNA using Phusion High-Fidelity polymerase (NEB, Beverly, MA) or by annealing complementary oligonucleotides (Venkova-Canova et al., 2006). The oriII plasmids in Fig. 5 contained a second origin (R6Koriγ) that was utilized during plasmid construction, where the host was CVC23, which supplies the cognate initiator (π protein).
Table 1.
Bacterial strains and plasmids used in this study 500
| Strains | Relevant characteristics | Source/Fig. |
|---|---|---|
| BR2846 | K-12 recA1 Δ (argF-lac)U169; Strain for cloning in pMLB1109 | (Simons et al., 1987) |
| BR8706 | =Stbl2 Δ(lac-proAB) Δ(araFGH) ΔaraEp PCP18-araE; araE under constitutive CP18 promoter | (Fekete & Chattoraj, 2005, Pal et al., 2005) |
| CVC23 | DH5(λpir); Strain for maintaining R6Koriγ plasmids. | M. Waldor |
| CVC72 | GM7589=MG1655 rph-1 lacU169 mutL459::Kan dam-13::Tn9; Dam− strain | M. Marinus |
| CVC73 | GM7576= MG1655 rph-1 lacU169 recA56 sr1300::Tn10; Dam+ strain | M. Marinus |
| CVC205 | V. cholerae El Tor N16961 lacZ::res::tet::res | (Pal et al., 2005) |
| CVC250 | CVC205 recA::Kan | (Pal et al., 2005) |
| DH5α | Strain for cloning | Invitrogen |
| Plasmids | ||
| pAS1 | PrctB (nt 1049–1133) in pMLB1109; ApR | Figs. 1, 2 |
| pDP309 | Same as pAS1, but PrctB (nt 256–1157) | (Pal et al., 2005) |
| pDP310 | Same as pAS1, but PrctB (nt 727–1157) | (Pal et al., 2005) |
| pDP332 | Same as pAS1, but PrctB (nt 547–1157) | (Pal et al., 2005) |
| pDP336 | Same as pAS1, but PrctB (nt 925–1157) | (Pal et al., 2005) |
| pMLB1109 | Source of promoter-less lacZ gene; pBR322ori; ApR | M. Berman |
| pSA5 | PrctB (nt 1049–1094) in pMLB1109; ApR | Fig. 1 |
| pTVC11 | rctB (nt 1118–3115) under PBAD control in pSC101ori; SpR | (Pal et al., 2005) |
| pTVC12 | PBAD in pSC101ori; SpR | (Pal et al., 2005) |
| pTVC25 | oriII (nt 441–1133) in R6Koriγ plasmid; ApR | (Venkova-Canova et al., 2006) |
| pTVC31 | Same as pTVC25, but oriII (nt 775–1133) | (Pal et al., 2005) |
| pTVC35 | pTVC31 lacking R6Koriγ (maintained in BR8706 + pTVC11) | (Venkova-Canova et al., 2006) |
| pTVC61 | pBR322ori. Digestion with EcoRV and HpaI leaves 300 bp flanks on either side of MCS; CmR | (Venkova-Canova et al., 2006) |
| pTVC126 | PrctA (nt 377-249) in pMLB1109; ApR | Fig. 2 |
| pTVC191 | pTVC61+ 39-mer from rctA (nt 176 – 214) | (Venkova-Canova & Chattoraj, 2011) |
| pTVC222 | pTVC243 + 39-mer (nt 449–487) | Fig. 3 |
| pTVC228 | 6×12-mers (nt 788–934) in pTVC243; CmR | (Venkova-Canova & Chattoraj, 2011) |
| pTVC234 | PrepA from P1 (nt 556–600) in pMLB1109 | (Venkova-Canova & Chattoraj, 2011) |
| pTVC243 | pTVC61 shorter by 400 bp. Digestion with EcoRV and HpaI leaves 100 bp flanks on either side of MCS; CmR | (Venkova-Canova & Chattoraj, 2011) |
| pTVC327 | pTVC243 + PrctB (same as in pAS1) | Fig. 3 and S2 |
| pTVC328 | pTVC243 + oriII (nt 1049–1094) | Fig. S2 |
| pTVC329 | pTVC222 with 5 bp deleted from the left side of the 39-mer linker region | (Venkova-Canova & Chattoraj, 2011) |
| pTVC330 | Same as pTVC329, but the deletion is of 10 bp | (Venkova-Canova & Chattoraj, 2011) |
| pTVC331 | pTVC222 with 5 bp deleted from the middle of the 39-mer linker region | (Venkova-Canova & Chattoraj, 2011) |
| pTVC332 | Same as pTVC331, but the deletion is of 10 bp | (Venkova-Canova & Chattoraj, 2011) |
| pTVC338 | pTVC243 + oriII (nt 1089–1118) | Fig. S2 |
| pTVC339 | pTVC243 + oriII (nt 1089–1132) | Fig. S2 |
| pTVC400 | pTVC243 + oriII (nt 1083–1124) | (Venkova-Canova & Chattoraj, 2011) |
| pTVC522 | pTVC61 + same oriII as in pTVC400 | Fig. 4 |
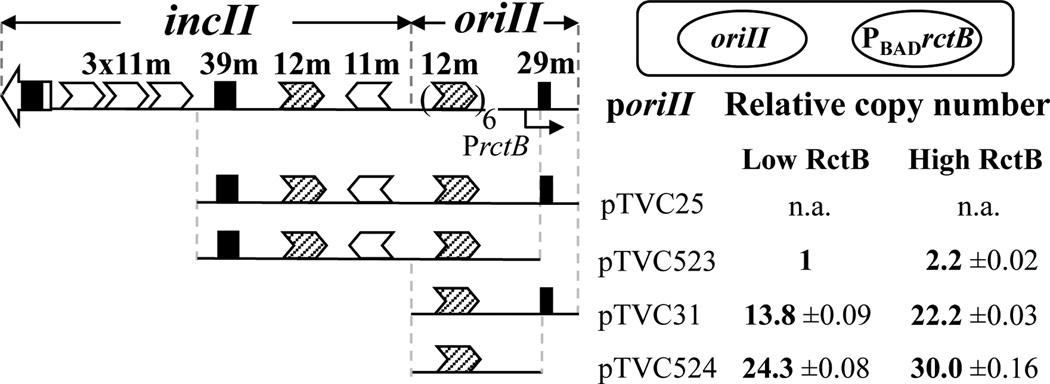
| pTVC523 | Same as pTVC25, but with deleted 29-mer (oriII = nt 441–1094) | Fig. 5 |
| pTVC524 | Same as pTVC31, but with deleted 29-mer (oriII = nt 775–1094) | Fig. 5 |
Fig. 5. The 29-mer, like the 39-mer, reduces replication of oriII-based plasmids in E. coli.
Four oriII-plasmids were tested for replication in the presence of an inducible source of RctB, as in Fig. 1 (cartoon on the top). Plasmids pTVC25 and pTVC523 are isogenic except for the presence of the 29-mer. They both have a part of the incII control region. Plasmids pTVC31 and pTVC524 are also isogenic except for the 29-mer but they lack incII totally. The oriII plasmid copy number was measured upon induction with 0.002% (Low RctB) and 0.2% (High RctB) arabinose, as described in Material and Methods. There was seven-fold more RctB at the higher inducer concentration, as was found earlier (Fig. S2 in Venkova-Canova & Chattoraj, 2011). The copy numbers shown are mean values from three cultures, each inoculated with separate single colonies. No transformants were obtained using pTVC25, hence its copy number is not shown (n.a.).
2.2 β-galactosidase assay
This was done from late log phase cultures in L broth (OD = 0.4–0.5) of E. coli strains, BR8706 (Fig. 1) and CVC72 -73 (Fig. 2), and of V. cholerae recA strain CVC250 (Fig. 1B), as described (Pal et al., 2005). Some of the cultures were simultaneously monitored for RctB induction by Western blotting (Fig. S1).
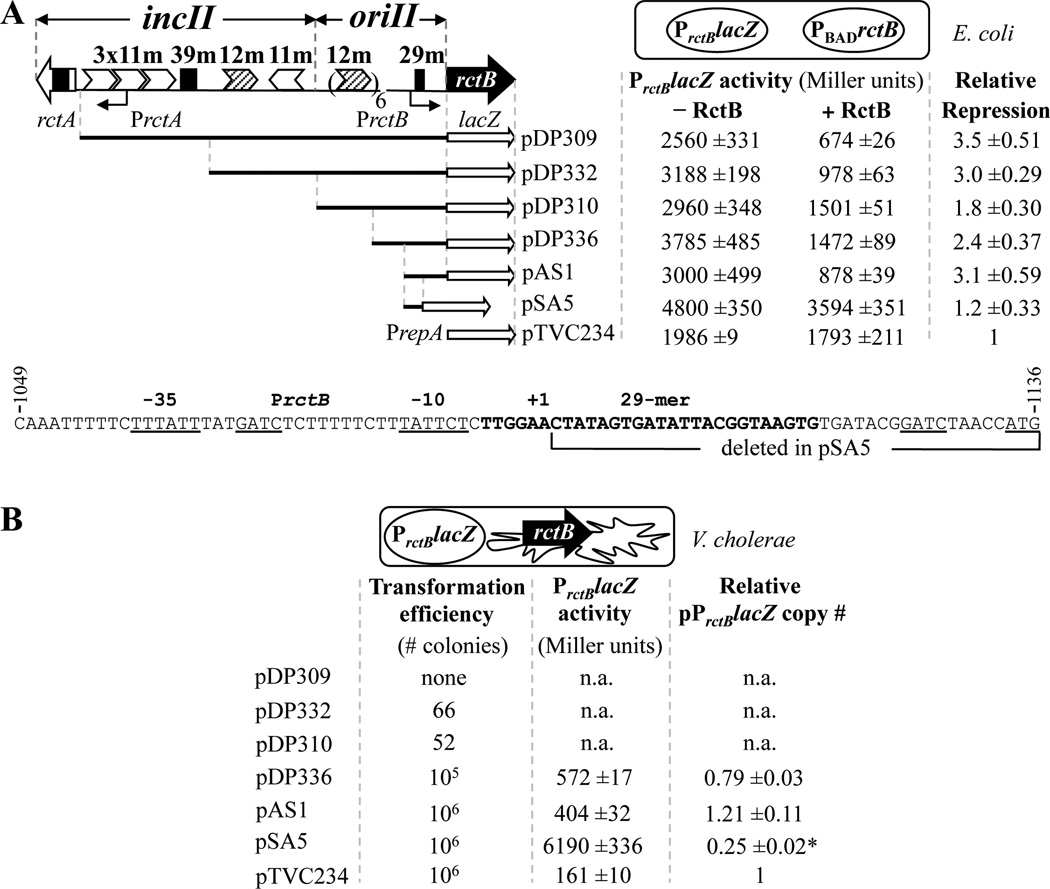
Fig. 1. A minimal promoter region that suffices for autorepression of rctB contains a 29-mer sequence.
A. Repression of PrctB by an inducible source of RctB in E. coli. A map of the replication region of V. cholerae chrII is shown at the top left. The region is divided into three segments: incII that regulates replication, oriII that is required in cis to initiate replication, and rctB (black arrow), the gene that codes for the initiator, RctB. RctB binds to two kinds of site: iterons, which are 11- or 12-mers (white or hatched arrowheads, respectively), and the 39-mers (black rectangles). The region has two promoters, PrctA and PrctB, the latter for transcribing rctB. A 29-mer, which has the conserved GC-rich direct repeats of 39-mers (boxed bases shown in Fig. 3) serves as the operator regulating PrctB, as shown below.
To identify the minimal region required for autorepression of rctB, the promoter with decreasing upstream sequences was fused to lacZ (white arrows) in a pBR322-derived vector. An identical construct (pTVC234) except where PrctB was replaced with a foreign promoter, PrepA, was used as the negative control. The autorepression was checked by supplying RctB from an inducible source, pTVC11, where rctB was under PBAD control (PBADrctB, cartoon on top right). 0.2% arabinose was used as the inducer. β-galactosidase activities were normalized with respect to the copy number of the relevant lacZ-plasmids (see Material and Methods). The values are mean ± one standard deviation from three independent measurements.
The sequence of the PrctB region present in pSA1 is shown at the bottom. The sequence coordinates are from the annotated chrII sequence at NCBI (NC_002506). -35 and -10 elements of PrctB are underlined. Also underlined are the two Dam methylation sites (GATC) and the starting ATG codon of rctB. +1 corresponds to the start position rctB mRNA. The 29-mer site is shown in bold. The region of pAS1 deleted in pSA5 (coordinates 1095–1136) is shown by a bracket.
B. Repression of PrctB by the natural source of RctB in V. cholerae. The same plasmids as in A were electroporated into CVC250 (V. cholerae recA mutant strain) and where colonies with normal size were obtained, β-galactosidase activity and copy number of the lacZ-carrying plasmids were measured. In the case of plasmids pDP309, -332 and -310, either transformants were not seen or they were bigger than normal size colonies (most likely these were revertants where the incII got deleted). The star indicates a slow-growing culture. The values are mean ± one standard deviation from three cultures inoculated with independent colonies.
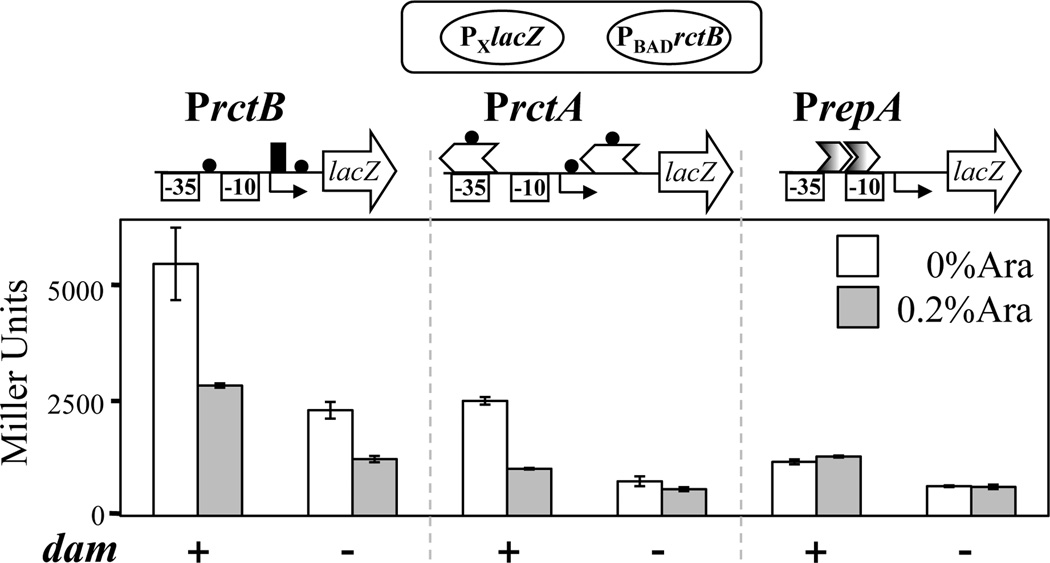
Fig. 2. Autorepression of rctB does not require methylation by Dam.
Repression by RctB was determined, as in Fig. 1, for three promoters, PrctB, PrctA and PrepA, present in plasmids pAS1, pTVC126 and pTV234, respectively. All promoter fragments contained the untranslated leader region and ended with their respective ATG start codon, which was fused to lacZ. The symbols are as in Fig. 1, except that GATC methylation sites are represented as black circles, and the -35 and -10 elements of the promoter are boxed. The bars show β galactosidase activity in Miller units, measured before (white bars) and after (grey bars) induction with 0.2% arabinose. The host cells either had the dam gene intact (+, CVC73) or inactivated (−, CVC72). The data are from three independent colonies for each of the fusions. The error bars here and elsewhere represent one standard deviation.
2.3 Plasmid copy-number measurements
The copy number was measured by supplying RctB from pTVC11, where rctB is expressed from an arabinose-inducible promoter, PBAD. E. coli BR8706 was used as a host, where the araE transporter gene is expressed constitutively from the chromosome. BR8706/pTVC11 cells were further transformed with oriII plasmids shown in Fig. 5. For Fig. S2, BR8706/pTVC11 cells were first transformed with a plasmid carrying the minimal oriII (pTVC35). Next, the BR8706/pTVC11/pTVC35 cells were made competent for further transformation with pBR322-derived plasmids carrying the 39-mer or different derivatives of the 29-mer. Plasmid copy number measurements were done exactly as described (Venkova-Canova & Chattoraj, 2011).
2.4 EMSA and Ligation Assay
These were done essentially as described (Venkova-Canova et al., 2006, Venkova-Canova & Chattoraj, 2011, Das & Chattoraj, 2004). The fragments used in Fig. 3 were PCR amplified from the plasmids identified in the figure legend. The fragments contained, in addition to the specific target sequences, 100 bp of non-specific sequences at both the flanks. The flanking sequences were identical in all cases. The fragments used in Fig. 4 were cut out by HpaI and EcoRV from pTVC228 and pTVC552 that created 100 bp flanks for 6×12-mers and 300 bp flanks for the 29-mer, respectively. The ends of the fragments were dephosphorylated using shrimp alkaline phosphatase, after which the fragments were gel purified. A total of 1–5 pmol ends of each fragment were labeled in 50 µl with 8.5 pmol [γ-32P] ATP (50 µCi/reaction, Perkin-Elmer, Waltham, MA) using 30 units of T4 polynucleotide kinase (NEB) for 30 min at 37°C. The enzyme was inactivated for 15 min at 65°C and the fragments were purified using Sephadex G-50 column (0.8 ml bed volume; Roche Diagnostics, Indianapolis, IN). The amount of labeled DNA was determined by TCA precipitation (Mallinckrodt Baker Inc., Phillipsburg, NJ) using salmon sperm DNA (20 µg; Invitrogen, Carlsbad, CA) as a carrier. The binding was done exactly as described (Venkova-Canova et al., 2006). In the experiments of Fig. 4, the binding buffer was supplemented with 4% PVA (polyvinyl alcohol, Sigma, St. Luis, MO), a molecular crowding agent. One half of the binding reaction was loaded onto a 5% polyacrylamide gel (National Diagnostics, Atlanta, Georgia), run at room temperature at 12 V cm−1 in TBE buffer (89mMTris/89mM boric acid/2.5 mM EDTA, pH 8.3). The gels were dried, and the band intensities were determined using a phosphoimager (FLA-5100, Fujifilm, Edison, NJ). The other half of the binding reaction was treated with 300 Units T4 Ligase (NEB) for 10 min at RT. The ligation reaction was stopped by equal volume of 2× stop solution (2× Bromphenol blue/Xylencyanol dye with additional 2% SDS and 20% Glycerol). The mixture was vortexed, heated at 65°C for 10 min, and spun for 5 min at 13,000 rpm. One fourth of the supernatant was loaded on a 5% polyacrylamide gel. The subsequent steps were exactly as described for EMSA gels.
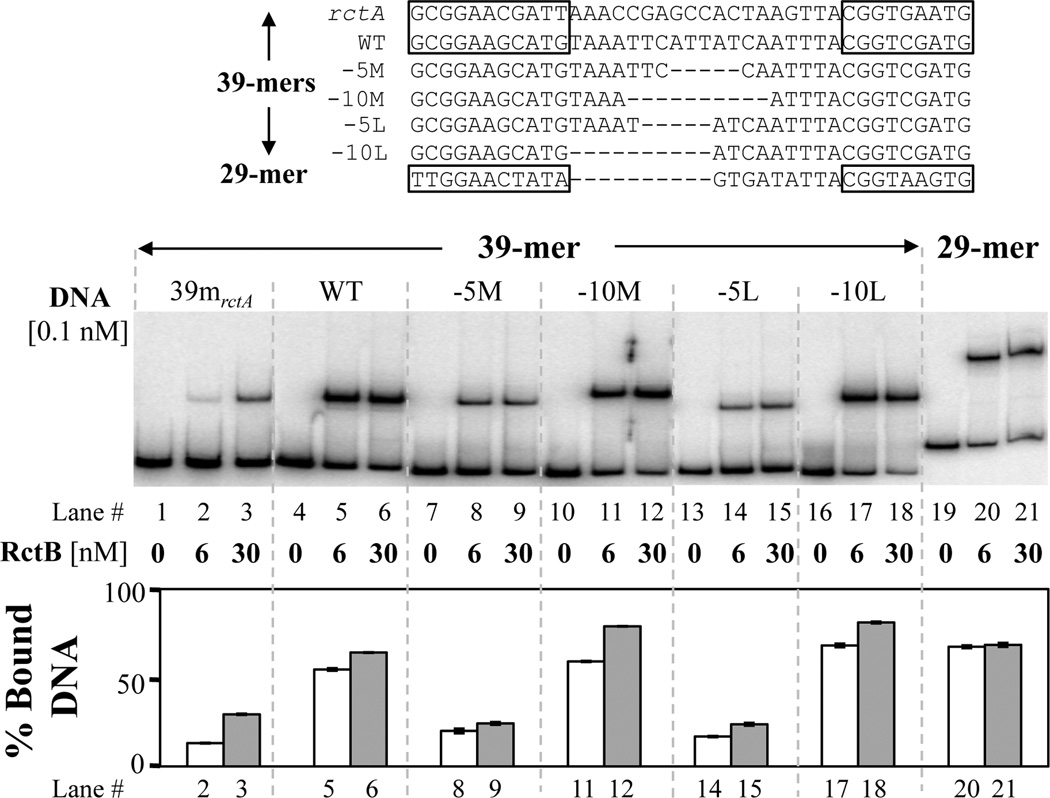
Fig. 3. The 29-mer, like the 39-mer, binds RctB in vitro.
The sequences of the 39-mers in rctA and in the middle of incII, and of several deletion derivatives of the latter are shown in the top, along with the sequence of the 29-mer. The 29-mer carrying fragment was bigger than the other fragments used here, as it contained more natural sequences flanking the site, exactly as present in pAS1 and shown on Fig. 1A (bottom sequence). To determine binding proficiency of these sequences to RctB, they were amplified by PCR (i.e. they were unmethylated) from the following plasmids: pTVC191, -222, -331, -332, -329, -330 and -327. The binding was monitored by EMSA (middle panel) at two different RctB concentrations. Percent binding ([intensity of the retarded band/combined intensities of free and retarded bands] × 100) at 6 nM and 30 nM are shown by white and grey bars, respectively. The error bars are from three independent measurements of band intensities from the same gel, which reflect the reproducibility of our measurements.
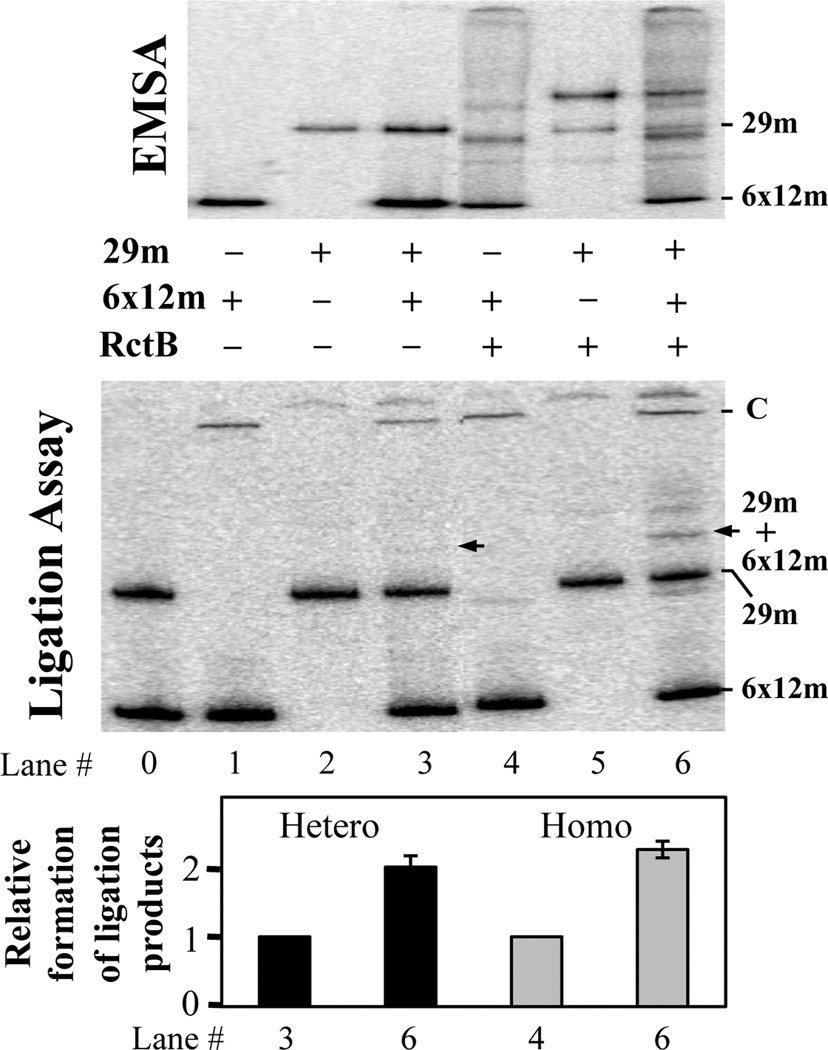
Fig. 4. The 29-mer, like the 39-mer, participates in handcuffing with iterons.
Two fragments (both at 0.02 nM), carrying the 29-mer in one and the 6×12-mers in the other, were analyzed by EMSA with 7 nM RctB, and subsequently, after addition of ligase to the binding reaction (Ligation Assay). The fragments were obtained by enzyme digestion from pTVC552 and pTVC228, respectively (see Material and Methods). Half of the binding reaction was loaded onto a 5% polyacrylamide gel (EMSA, top panel). The remaining half was treated with ligase to record coupling rate of the fragments. The products of ligation, after deproteinization, were analyzed also in a 5% polyacrylamide gel (Ligation Assay, middle panel). The sample in lane 0 did not contain ligase. C stands for circular products of ligation, which migrate poorly in polyacrylamide gels. The DNA species representing a heterodimer of 29-mer and a 6×12-mer fragments is indicated by arrows. The intensities of this band before (lane 3) and after (lane 6) addition of RctB, are shown by black bars on the graph below. The intensities of the self-ligated products of the 6×12-mer fragments (Homo), are shown by grey bars after excluding the intensity of the heterodimer band and the 29-mer bands from lane 6. Note that self-ligation of the 29-mer fragment is not enhanced by RctB. Lanes 0 and 1 were used for background correction. The error bars are from measurement of three independent gels.
3. Results
3.1 A 29-mer site suffices for autorepression of rctB
The promoter of rctB, PrctB, was defined earlier by mapping the start site of rctB message (Pal et al., 2005, Egan et al., 2006). These studies also showed that the promoter is autorepressed. To identify the binding site(s) of RctB relevant to autorepression, the promoter upstream region was gradually shortened (Fig. 1). The promoter activity was measured in E. coli as before, using lacZ as the reporter gene, without and with RctB induction from pTVC11, in which the initiator gene is under the control of an arabinose-inducible promoter, PBAD. The promoter of plasmid P1 initiator, PrepA, present in pTVC234, served as the negative control. The DNA upstream of PrctB did not affect significantly the promoter activity in the absence of RctB induction. Upon RctB induction (Fig. S1), the promoter activities decreased up to about four-fold, and the level of decrease depended upon the extent of the upstream region present. The reason for variable repression is likely to be due to the presence of several RctB binding sites in the upstream sequences that could influence the repression indirectly by titrating the initiator or directly by coupling interactions with the promoter region (discussed in section 3.4). Although a clearer interpretation of these results must await further analysis, it appears that the shortest region tested for promoter regulation (as in pAS1) showed not only optimal promoter activity but also near maximal repression (3.1 fold). This region will be referred hereafter as minimal PrctB.
RctB binds to multiple sites that are of two kinds, the iterons (11- and 12-mers) and the 39-mers (Fig. 1A, top map) (Venkova-Canova & Chattoraj, 2011). For efficient binding, the iterons need to be methylated at the adenine residues of GATC sequences, which are found internal to all the iterons. Inspection of the chrII region present in pAS1 revealed two GATC sites but the homology to iterons did not extend beyond those four bases. However, a 29-mer site was found within the PrctB that was detected earlier by searching for sequences homologous to the 39-mers (Venkova-Canova & Chattoraj, 2011). The 29-mer lacks the first 10 bp of the AT-rich linker between the two conserved GC-rich direct repeats of the 39-mers (hyphens; Fig. 3 top). The 29-mer covers the − 4 to +25 bp of the template DNA for rctB transcription, where +1 corresponds to the start site of rctB mRNA (Fig. 1A, bottom). The 29-mer appears to be ideally situated to serve as an operator in that bound RctB should interfere with RNA polymerase binding and transcription initiation.
In an assay for replication driven by oriII, the 29-mer was shown earlier to be active in RctB titration, indicating that the site was able to bind the initiator in vivo (Venkova-Canova & Chattoraj, 2011). The fragment used had six extra bases that are naturally present, flanking the 29-mer. Without the flanks, the titration activity was less (Fig. S2; fragments 4 vs. 5). Deleting a good part of the 29-mer abrogated the titration activity entirely (Fig. S2; fragment 2). Same deletion in pSA5 did not affect the promoter activity but abolished the capacity for autorepression. It appears that a few extra flanking bases can make the 29-mer more efficient in RctB titration.
The 29-mer was effective as an operator in the native host also. Although the amount of RctB in V. cholerae was ~2 fold less than that present under the inducible conditions used in E. coli (Fig. S1), PrctB activity from pAS1 was significantly lower than the activity from pSA5 (Fig. 1B). In summary, it appears that the minimal promoter, present in pAS1, is necessary and sufficient for rctB expression and autorepression, where the 29-mer serves as the operator, in both E. coli and V. cholerae.
3.2. rctB autorepression does not require methylation by Dam
As stated above, the minimal promoter, in addition to the 29-mer, contains two GATC sites (Fig.1A, bottom). These two could be part of putative iterons; although iteron bases show some homology over a stretch of 11 to 12 bp, full conservation is found only in six consecutive positions that include GATC. Since the iterons require methylation for efficient RctB binding, we hypothesized that if the putative iterons of the minimal promoter were required for autorepression, it might be compromised in dam mutant cells. To test the hypothesis, we used PrctA as positive control, which carries two natural 11-mers, one overlapping with its -35 element; RctB binding and repression of this promoter is therefore expected to be dependent on Dam. As a negative control, we used PrepA, which is devoid of GATC sequences. The promoter activities were measured exactly as in Fig. 1. In wild-type (dam+) cells, PrctA and PrctB were repressed upon induction of RctB, as opposed to the negative control PrepA, whose activity was unchanged by the induction (Fig. 2). This indicates that minimal PrctB region includes a binding site of RctB. In dam mutant cells, activity of all the three promoters tested was down, even without RctB induction. This could be due to altered physiology of the mutant cells and/or the altered DNA structure (Lobner-Olesen et al., 2005, Polaczek et al., 1998). Induction of RctB in the dam mutant not only failed to repress PrepA but also PrctA, consistent with the requirement of methylation for RctB binding to iterons (Fig.2). In contrast, PrctB remained sensitive to RctB induction; the promoter activity dropped by ~50%, similar to what was found in dam+ cells. Thus, the only identifiable RctB binding site within the minimal PrctB region, the 29-mer, appears to be the operator for rctB autorepression, and it functions without requiring Dam.
3.3 The 29-mer is a binding site of RctB in vitro
To confirm RctB binding to the 29-mer in vitro, EMSA was performed with pure protein. As controls, the 39-mers from rctA and from the middle of incII with its various deletion derivatives were used (Fig. 3, top). The alterations were made in the linker region of the 39-mer by deleting either 10 bp to mimic the 29-mer, or five bp, which served as a control. The 29-mer bound RctB with affinity similar to that of the 39-mer. The binding was also comparably efficient when the 39-mer was shortened by 10 bp at two different places of the linker (Fig. 3, bottom). The binding was reduced when five bp was deleted. These results suggest that the 29-mer is a natural example of a 39-mer deleted for 10 bp, and the deletion does not affect the binding affinity of the sites.
The better binders were also more effective in reducing the copy number of a plasmid driven by oriII in vivo (Venkova-Canova & Chattoraj, 2011). In this assay, the intact 39-mer was most potent in reducing the copy number, followed by the 39-mer from rctA and the three 29-mers, the natural and the two artificially made ones, which were ~50% as effective. Thus, although the 29-mers and the 39-mer bound equally well in vitro, their activities in vivo were not the same. Since in spite of similar binding the control activity of the deleted sites were reduced by 50%, the results suggest that either EMSA overestimated the binding affinity of the 29-mers or the structure of RctB bound to 29-mer and 39-mer differs enough to affect the function of the complexes.
3.4 The 29-mer, like the 39-mer, participates in handcuffing with iterons
It is known that RctB, like the plasmid initiators, not only binds its target sites but also couples them, a reaction commonly referred to as handcuffing (Venkova-Canova & Chattoraj, 2011). RctB can couple iterons, iterons with 39-mers (hetero-handcuffing) but not the 39-mers. The 39-mers, however, improved handcuffing of iterons with themselves. Most likely, RctB binding to 39-mers changes the conformation of the initiator such that it becomes more proficient in handcuffing of iterons. The handcuffing is believed to engage the iterons of the origin in a way that impairs origin function. Because of the sequence similarity between the 29-mer and the 39-mers, we tested whether the 29-mer could also participate in handcuffing, as do the 39-mers. EMSA, followed by ligation assay were performed exactly as for the 39-mer (Venkova-Canova & Chattoraj, 2011). First, binding was tested by EMSA (Fig. 4, top). The fragments carrying the 29-mer and the origin 6×12-mers bound RctB to a comparable extent, whether the fragments were tested separately (lanes 4 and 5) or together (lane 6). After treatment of the binding reaction with ligase, the 29-mer showed little difference in ligated products in the absence or the presence of RctB (lanes 2 and 5, Fig. 4 Ligation assay). This result is consistent with our earlier result showing that the 39-mers do not handcuff with themselves (Venkova-Canova & Chattoraj, 2011). In contrast, the 6×12-mers clearly showed more ligated products upon addition of RctB, confirming the earlier finding that RctB can handcuff iterons (lanes 1 and 4, Fig. 4 Ligation assay). When the two fragments were together, the band representing ligation of the 29-mer fragment with the 6×12-mer fragment was enhanced in the presence of RctB, indicating that the 29-mer can hetero-handcuff with the iterons (Fig. 4, arrows, and the black bars in graph below). An increase in the ligated products among the iteron fragments themselves in presence of the 29-mer was also evident (Fig. 4, lane 6 in the graph below). In these experiments, the handcuffing of iterons needs to be kept low since homo-handcuffing of iterons dominates hetero-handcuffing, making the latter reaction harder to detect. In conclusion, it appears that the handcuffing properties of the 29-mer are quite similar to those of the 39-mers.
3.5 The 29-mer reduces replication of oriII-based plasmids in E. coli
The similarity of the handcuffing properties of the 29-mer with those of the 39-mers in vitro suggests that the 29-mer could be controlling chrII replication in vivo, as do the 39-mers. This was tested by measuring the copy number of oriII plasmids with and without the 29-mer in cis. A two-plasmid system was used in E. coli, where one had oriII and the other provided RctB in trans from pTVC11. The use of a surrogate host has the advantage that the issue of incompatibility between the plasmid and chromosomal origins does not arise (Venkova-Canova & Chattoraj, 2011, Egan & Waldor, 2003, Yamaichi et al., 2011). In these experiments, oriII plasmids were electroporated into an E. coli that already harbored the RctB plasmid. No colonies were seen when pTVC25 was used for transformation, most probably because the 29-mer acted as a negative regulator on top of the strong negative regulator, the 39-mer. Transformants were obtained with an isogenic plasmid pTVC523 lacking the 29-mer. The presence of the plasmid in the transformants was confirmed by measuring its copy number (Fig. 5). The results were similar with the plasmid pair pTVC31 and pTVC524, with and without the 29-mer, respectively. Both these plasmids replicated with high copy numbers because they lack the negative control locus, incII, entirely. Even in this high-copy-number plasmid set, the one without the 29-mer had the higher copy number. This could be due to several reasons: (i) lack of titration of RctB by the 29-mer, which will increase initiator availability for replication initiation; (ii) lack of interactions between 29-mer and the origin iterons (hetero-handcuffing) which will improve the origin function; and (iii) activation of the origin by increased activity of PrctB in the absence of the operator site (transcriptional activation (Baker & Kornberg, 1988)). We conclude that the 29-mer is capable of inhibiting replication even in the presence of other regulators of incII. This regulatory role is in addition to the role of the 29-mer in autorepression, since in the present experiment RctB was supplied from a foreign promoter (PBAD) in trans that is not under RctB control.
4. Discussion
Autorepression is an efficient mechanism to maintain gene products within narrow limits and to dampen out the effect of changes in gene-dosage (Becskei & Serrano, 2000, Simpson et al., 2003). Many bacterial and plasmid initiators are autorepressed at the transcriptional level (Das et al., 2005, Atlung et al., 1985). This is easily rationalized since initiators are essential for replication and often the rate-limiting component of replication; therefore maintaining their concentration within narrow limits becomes necessary to maintain the rate of replication.
In the case of plasmids of the iteron-family, transcriptional autorepression is the main mechanism that limits initiator synthesis (Paulsson & Chattoraj, 2006). An exception has been found in plasmid RK2, where the initiator gene is regulated by the products of three other plasmid genes, korA, korB and kilB1 (Kolatka et al., 2010). There is a post-translational mechanism of autorepression as well, which operates by initiator dimerization (Paulsson & Chattoraj, 2006). In iteron-based plasmids, initiator dimers are inactive since only monomers bind iterons. The role of dimerization in dampening initiator monomer fluctuation has been deduced from sound theoretical considerations, and its contribution to maintaining monomers within narrow limits is predicted to be similar to that of transcriptional autorepression. In either of the mechanisms, the autorepression is not known to depend on the cell cycle.
The chrII iterons require full methylation for binding to the initiator, whereas plasmid iterons do not (Demarre & Chattoraj, 2010). ChrII also replicates at a particular time of the cell cycle whereas initiation of plasmid replication is distributed throughout the cell cycle. The involvement of methylation allows controlling the iteron activity in the cell cycle, since they become inactive following replication, and stay inactive for about two-thirds of the cell cycle. In this context, use of a 29-mer as the operator should help maintain the initiator in a cell-cycle independent fashion, since the site lacks the sequences for methylation. Finally, the methylation-independent regulation of the initiator expression should allow restricting the amount of the newly-synthesized RctB during the hemi-methylated period, which would prevent over-loading of cells with RctB when oriII becomes methylated again. This should be beneficial for the once-per-cell-cycle replication initiation.
The chrII replication initiation rate increases in response to increases in RctB concentration; in contrast, the copy number of iteron-carrying plasmids hardly changes when initiator concentration is increased (Das et al., 2005, Pal et al., 2005, Duigou et al., 2006, Pal & Chattoraj, 1988, McEachern et al., 1989). This suggests that there might be a greater demand to maintain RctB concentration within narrower limits than what might suffice for the plasmid initiators. In chrII, the mechanism of transcription repression itself might be more complex, involving sequences other than the 29-mer, as suggested by the results of Fig. 1, where sequences upstream of PrctB could influence the magnitude of autorepression. The influence could also vary during the cell cycle, since the iterons remain active only a part of the cell cycle (Demarre & Chattoraj, 2010). There could also be mechanisms other than transcription autorepression to make the regulation of RctB concentration more stringent.
The 29-mer, like the 39-mers, was able to inhibit oriII-based plasmid replication (Fig. 5). The site thus has a dual role in chrII replication. The role as transcription operator is perhaps more important since it is required to control the rate-limiting component of replication – the RctB concentration. The role as a replication inhibitor should be relatively minor since the 29-mer is one of several other regulatory sites involved in controlling chrII replication. Also, compared to the 39-mers, the 29-mer is less potent in inhibiting oriII-based plasmid replication (Venkova-Canova & Chattoraj, 2011). It is not clear whether there is any significance of having a 29-mer as the operator and 39-mers as purely replication inhibitors. To this end, we note that autorepression could also be achieved equally efficiently when the 29-mer was replaced with the 39-mer (data not shown). In any event, the ability of RctB to recognize 39-mer like sites when the linker sequences were changed from normally 19 to either nine or 29 bp shows the inherent adaptability of the RctB structure. Since not all binding proficient sites were functional in vivo, this suggests that the sites not merely titrate initiators but play more active role in replication, such as handcuffing. Here we have shown that the 29-mer, like the 39-mer, was capable of promoting self-ligation of iteron fragments in trans most likely by remodeling RctB. Further understanding of these results calls for more structural information of the complexes.
Highlights.
The replication initiator of V. cholerae chromosome II is limiting for replication.
The autorepression site for the initiator gene expression is identified.
The same site additionally serves as a negative regulator of replication.
Supplementary Material
Acknowledgements
We thank Arnab Sarkar and Souvanik Adhya, for plasmid construction. We are grateful to Michael Yarmolinsky for a critical review of an earlier draft of this paper. This work was supported by the Intramural Research Program, Center for Cancer Research, National Cancer Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Atlung T, Clausen ES, Hansen FG. Autoregulation of the dnaA gene of Escherichia coli K12. Mol Gen Genet. 1985;200:442–450. doi: 10.1007/BF00425729. [DOI] [PubMed] [Google Scholar]
- Baker TA, Kornberg A. Transcriptional activation of initiation of replication from the E. coli chromosomal origin: an RNA-DNA hybrid near oriC. Cell. 1988;55:113–123. doi: 10.1016/0092-8674(88)90014-1. [DOI] [PubMed] [Google Scholar]
- Becskei A, Serrano L. Engineering stability in gene networks by autoregulation. Nature. 2000;405:590–593. doi: 10.1038/35014651. [DOI] [PubMed] [Google Scholar]
- Chattoraj DK. Control of plasmid DNA replication by iterons: no longer paradoxical. Mol Microbiol. 2000;37:467–476. doi: 10.1046/j.1365-2958.2000.01986.x. [DOI] [PubMed] [Google Scholar]
- Das N, Chattoraj DK. Origin pairing ('handcuffing') and unpairing in the control of P1 plasmid replication. Mol Microbiol. 2004;54:836–849. doi: 10.1111/j.1365-2958.2004.04322.x. [DOI] [PubMed] [Google Scholar]
- Das N, Valjavec-Gratian M, Basuray AN, Fekete RA, Papp PP, Paulsson J, Chattoraj DK. Multiple homeostatic mechanisms in the control of P1 plasmid replication. Proc Natl Acad Sci U S A. 2005;102:2856–2861. doi: 10.1073/pnas.0409790102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarre G, Chattoraj DK. DNA adenine methylation is required to replicate both Vibrio cholerae chromosomes once per cell cycle. PLoS Genet. 2010;6:e1000939. doi: 10.1371/journal.pgen.1000939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duigou S, Knudsen KG, Skovgaard O, Egan ES, Lobner-Olesen A, Waldor MK. Independent Control of Replication Initiation of the Two Vibrio cholerae Chromosomes by DnaA and RctB. J Bacteriol. 2006;188:6419–6424. doi: 10.1128/JB.00565-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan ES, Duigou S, Waldor MK. Autorepression of RctB, an initiator of Vibrio cholerae chromosome II replication. J Bacteriol. 2006;188:789–793. doi: 10.1128/JB.188.2.789-793.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan ES, Fogel MA, Waldor MK. Divided genomes: negotiating the cell cycle in prokaryotes with multiple chromosomes. Mol Microbiol. 2005;56:1129–1138. doi: 10.1111/j.1365-2958.2005.04622.x. [DOI] [PubMed] [Google Scholar]
- Egan ES, Waldor MK. Distinct replication requirements for the two Vibrio cholerae chromosomes. Cell. 2003;114:521–530. doi: 10.1016/s0092-8674(03)00611-1. [DOI] [PubMed] [Google Scholar]
- Fekete RA, Chattoraj DK. A cis-acting sequence involved in chromosome segregation in Escherichia coli. Mol Microbiol. 2005;55:175–183. doi: 10.1111/j.1365-2958.2004.04392.x. [DOI] [PubMed] [Google Scholar]
- Fogel MA, Waldor MK. A dynamic, mitotic-like mechanism for bacterial chromosome segregation. Genes Dev. 2006;20:3269–3282. doi: 10.1101/gad.1496506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PW, Lower RP, Kim NK, Young JP. Introducing the bacterial 'chromid': not a chromosome, not a plasmid. Trends Microbiol. 2010;18:141–148. doi: 10.1016/j.tim.2009.12.010. [DOI] [PubMed] [Google Scholar]
- Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Umayam L, Gill SR, Nelson KE, Read TD, Tettelin H, Richardson D, Ermolaeva MD, Vamathevan J, Bass S, Qin H, Dragoi I, Sellers P, McDonald L, Utterback T, Fleishmann RD, Nierman WC, White O, Salzberg SL, Smith HO, Colwell RR, Mekalanos JJ, Venter JC, Fraser CM. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julio SM, Heithoff DM, Provenzano D, Klose KE, Sinsheimer RL, Low DA, Mahan MJ. DNA adenine methylase is essential for viability and plays a role in the pathogenesis of Yersinia pseudotuberculosis and Vibrio cholerae. Infect Immun. 2001;69:7610–7615. doi: 10.1128/IAI.69.12.7610-7615.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolatka K, Kubik S, Rajewska M, Konieczny I. Replication and partitioning of the broad-host-range plasmid RK2. Plasmid. 2010;64:119–134. doi: 10.1016/j.plasmid.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Lobner-Olesen A, Skovgaard O, Marinus MG. Dam methylation: coordinating cellular processes. Curr Opin Microbiol. 2005;8:154–160. doi: 10.1016/j.mib.2005.02.009. [DOI] [PubMed] [Google Scholar]
- McEachern MJ, Bott MA, Tooker PA, Helinski DR. Negative control of plasmid R6K replication: possible role of intermolecular coupling of replication origins. Proc Natl Acad Sci U S A. 1989;86:7942–7946. doi: 10.1073/pnas.86.20.7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal D, Venkova-Canova T, Srivastava P, Chattoraj DK. Multipartite regulation of rctB the replication initiator gene of Vibrio cholerae chromosome II. J Bacteriol. 2005;187:7167–7175. doi: 10.1128/JB.187.21.7167-7175.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal SK, Chattoraj DK. P1 plasmid replication: initiator sequestration is inadequate to explain control by initiator-binding sites. J Bacteriol. 1988;170:3554–3560. doi: 10.1128/jb.170.8.3554-3560.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsson J, Chattoraj DK. Origin inactivation in bacterial DNA replication control. Mol Microbiol. 2006;61:9–15. doi: 10.1111/j.1365-2958.2006.05229.x. [DOI] [PubMed] [Google Scholar]
- Polaczek P, Kwan K, Campbell JL. GATC motifs may alter the conformation of DNA depending on sequence context and N6-adenine methylation status: possible implications for DNA-protein recognition. Mol Gen Genet. 1998;258:488–493. doi: 10.1007/s004380050759. [DOI] [PubMed] [Google Scholar]
- Rasmussen T, Jensen RB, Skovgaard O. The two chromosomes of Vibrio cholerae are initiated at different time points in the cell cycle. EMBO J. 2007;26:3124–3131. doi: 10.1038/sj.emboj.7601747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons RW, Housman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- Simpson ML, Cox CD, Sayler GS. Frequency domain analysis of noise in autoregulated gene circuits. Proc Natl Acad Sci U S A. 2003;100:4551–4556. doi: 10.1073/pnas.0736140100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Val ME, Kennedy SP, El Karoui M, Bonne L, Chevalier F, Barre FX. FtsK-dependent dimer resolution on multiple chromosomes in the pathogen Vibrio cholerae. PLoS Genet. 2008;4:e1000201. doi: 10.1371/journal.pgen.1000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkova-Canova T, Chattoraj DK. Transition from a plasmid to a chromosomal mode of replication entails additional regulators. Proc Natl Acad Sci U S A. 2011;108:6199–6204. doi: 10.1073/pnas.1013244108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkova-Canova T, Srivastava P, Chattoraj DK. Transcriptional inactivation of a regulatory site for replication of Vibrio cholerae chromosome II. Proc Natl Acad Sci U S A. 2006;103:12051–12056. doi: 10.1073/pnas.0605120103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaichi Y, Gerding MA, Davis BM, Waldor MK. Regulatory cross-talk links Vibrio cholerae chromosome II replication and segregation. PLoS Genet. 2011;7:e1002189. doi: 10.1371/journal.pgen.1002189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.