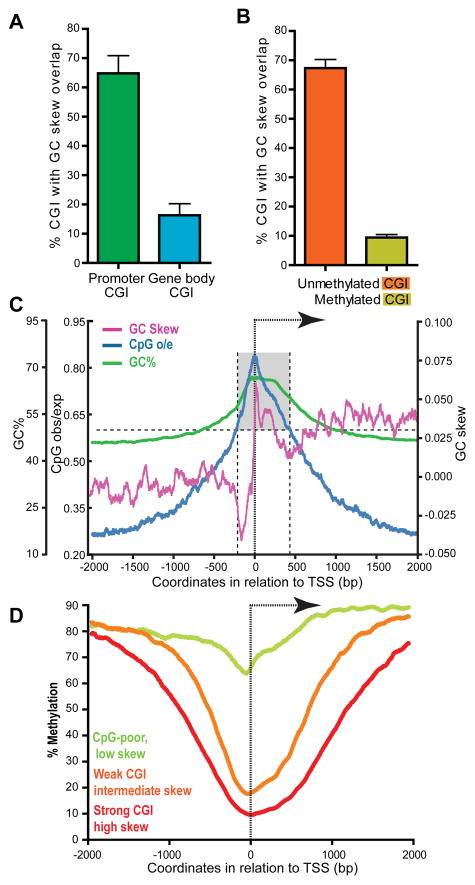
Figure 6. R-loop formation potential is predictive of the unmethylated status of CGI promoters.
A Percent of promoter CGIs (n=13,636) and gene body CGIs (n=4,598) with GC skew overlap. B Percent of unmethylated (n=1,785) and methylated (n=1,594) CGIs showing GC skew overlap (Methylation data from the hESC dataset from (Straussman et al., 2009)). Unmethylated CGIs showed a methylation score <−0.8 while methylated CGIs had a score > 1.3 in both hESC cell lines). In both panels, the overlap was calculated for each chromosome and the average and standard deviation for the genome are shown. C Metagene analysis of a subset of 4,528 promoter CGIs lacking strong GC skew. Symbols and analysis are as described for Figure 1C. D Aggregate CpG methylation levels around the TSS of genes corresponding respectively to strong CGIs characterized by high GC skew (n=7,820, red), weak CGIs characterized by intermediate GC skew (n=4,526, orange), and “CpG-poor” promoters characterized by little to no skew (n=7,570, green). The DNA methylation data was from (Laurent et al., 2010).