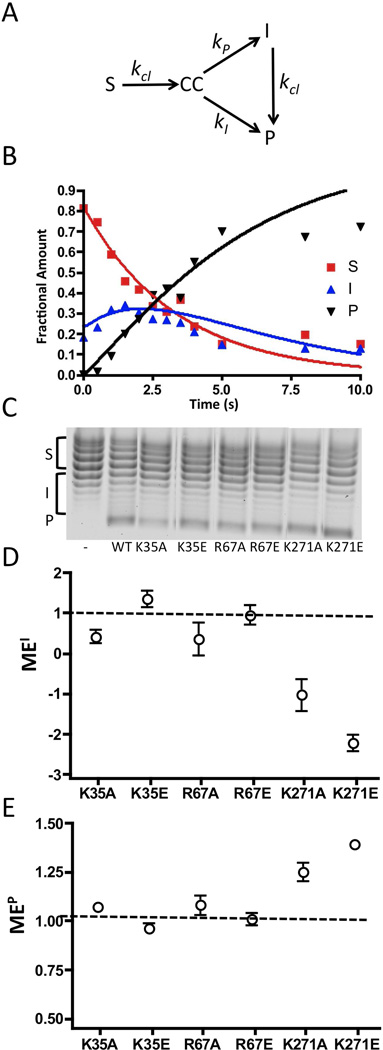
Figure 7.
Supercoil unwinding measurements. (A) Kinetic model for analysis of supercoil unwinding (see Supplemental Materials), and (B) the time course for supercoil unwinding by wild-type vTopo. The fraction of S, I and P at each time point were modeled using the numerical integration program Dynafit (16). The rates for S→CC, CC→I, CC→P and I→P are kcl, kI, kP, and kcl respectively. The fitted kinetic parameters for the wild-type enzyme were kcl = 0.3 s−1, kI = 5.0 s−1 kP = 2.3 s−1 (see Supplemental Materials for an analysis of K271A). The ratio kI/kP = 2 is a measure of how much of the covalent complex generated from rate limiting cleavage of S partitions to I as opposed to P. Thus, this ratio is a quantitative measure of processivity of supercoil unwinding that is analogous to the simple processivity factor (eq 4). (C) Representative chloroquine gel analysis for supercoil unwinding by wild-type and mutant topoisomerases as indicated. In this system, negatively supercoiled substrate topoisomers (S) migrate at the top of the gel and relaxed products (P) migrate at the bottom. Each reaction was terminated after 18 ± 2% of substrate had been consumed (see text). The left most lane is the pUC19 substrate in the absence of topoisomerase. The bands corresponding to S, I and P are shown by brackets along side the gel image (see Methods). (E) Mutational effect on the processivity factor (Pf) based on accumulation of intermediates MEI (see text). (E) Mutational effect on the processivity factor (Pf) based on appearance of product MEP (see text). The dashed line at one indicates no mutational effect.