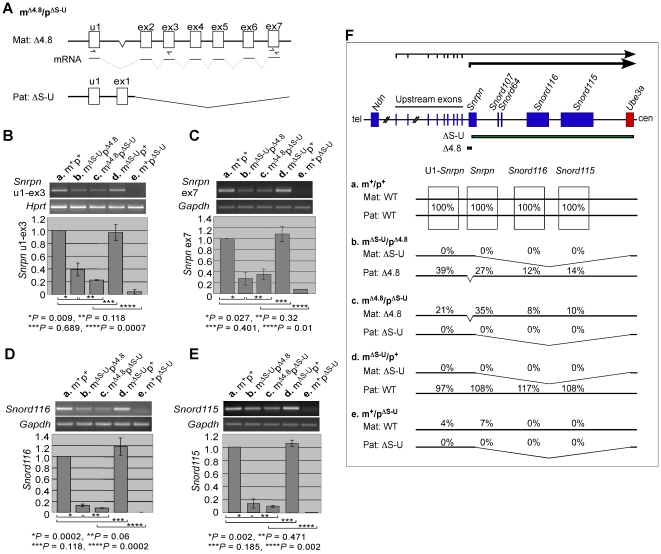
Figure 1. Expression analysis of Snrpn, Snrod116, and Snord115 in mice carrying the Δ4.8 mutation and/or the ΔS-U mutation.
(A) Genomic structure of the maternal Δ4.8 allele and the paternal ΔS-U allele in the mΔ4.8pΔS-U mice. The Δ4.8 mutation removes exon 1 of Snrpn. Snrpn is still able to transcribe from the upstream exons splicing to Snrpn exon 2. The relative positions of the primers specific for upstream exon 1 and exon 3 (u1-ex3) and for the downstream exon 7 (ex7) designed for RT-PCR and qRT-PCR are indicated (half-arrows). The ΔS-U mutation removes Snrpn from exon 2 to exon 10. (B–E) The Snrpn u1-ex3 (B), Snrpn exon 7 (C), Snrod116 (D), and Snord115 (E) transcripts were analyzed by RT-PCR (top) and quantitative RT-PCR (bottom). Total RNA was isolated from brains of wild-type mice (a, m+p+) (n = 5), mice inheriting the ΔS-U mutation maternally and the Δ4.8 mutation paternally (b, mΔS-UpΔ4.8) (n = 5), mice inheriting the Δ4.8 mutation maternally and the ΔS-U mutation paternally (c, mΔ4.8pΔS-U) (n = 5), mice with only the maternally inherited ΔS-U mutation (d, mΔS-Up+) (n = 5), and mice with only the paternally inherited ΔS-U mutation (e, m+pΔS-U) (n = 5). RT-PCR analyses were performed using 2.0 µg total RNA (top). For quantitative RT-PCR, the levels of gene expression from wild-type mice were set as 1 (bottom). Transcripts of Hprt were amplified as an endogenous control for the Snrpn u1-ex3 transcripts, since their sizes were similar. Transcripts of Gapdh were amplified as an endogenous control for the Snrpn exon 7, Snrod116, and Snord115 transcripts. RT-PCR products: Snrpn u1-ex3, 295 bp; Hprt, 266 bp; Snrpn ex7, 171 bp; Snrod116, 98 bp; Snrod115, 79 bp; Gapdh, 97 bp. (F) Schematic representation of the mouse PWS/AS domain (top) and summary of gene expression in mice of the five different genotypes (bottom, a–e). The Snrpn sense/Ube3a antisense transcripts initiated from Snrpn exon 1 with the major promoter activity and from Snrpn upstream exons with weaker promoter activity are marked as bold and thin arrows, respectively. SnoRNAs are encoded within these large Snrpn sense/Ube3a antisense transcripts derived from both Snrpn major and upstream exon promoters. Snord116 and Snord115 are multiple copy gene clusters. The centromeric (cen) and the telomeric (tel) positions are indicated. Paternally and maternally expressed genes are marked as blue and red boxes, respectively. ΔS-U indicates a large deletion from Snrpn exon 2 to Ube3a. Δ4.8 indicates a 4.8-kb deletion at Snrpn exon 1. The levels of the Snrpn u1-ex3, Snrpn exon 7, Snrod116, and Snord115 transcripts from wild-type mice were set as 100%. Mat, maternal chromosome; Pat, paternal chromosome.