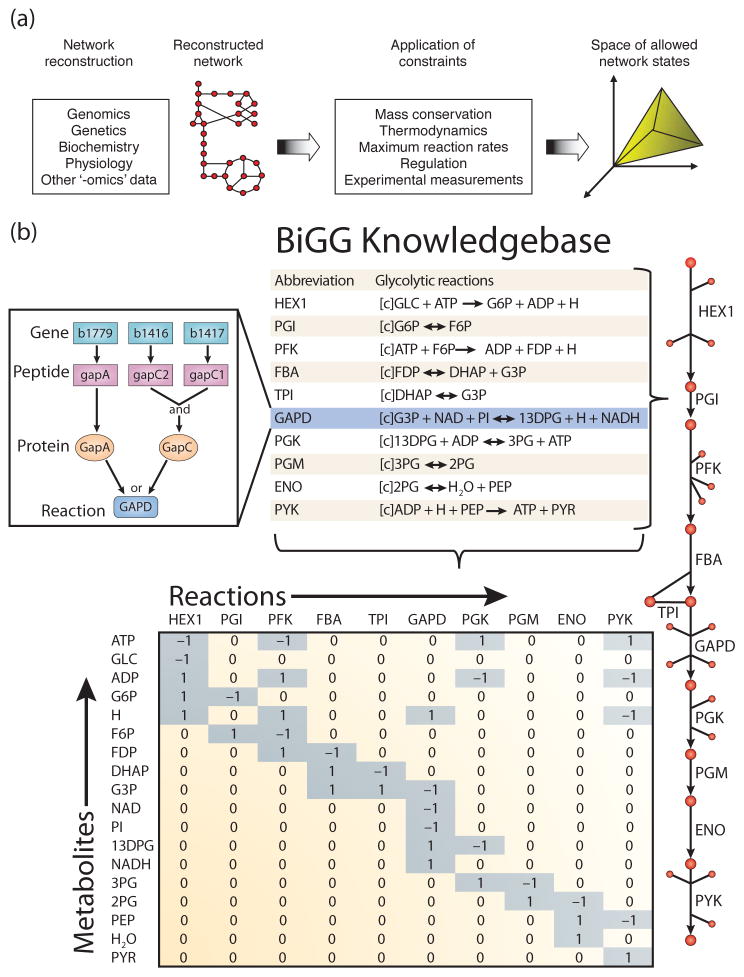
Figure 1. The philosophy of COnstraints-Based Reconstruction and Analysis.
(a) COnstraints-Based Reconstruction and Analysis (COBRA) of biological networks involves the creation of network models from a variety of biological data sources. The capabilities of the model are then assessed in the context of physical, chemical, regulatory, and omics constraints (Reproduced from Becker et al.17 with permission). (b) COBRA models are often derived from BiGG knowledgebases which are essentially 2-D annotations of the genome that relate metabolic activity to genomic loci. (left inset) In Escherichia coli, the glyceraldehyde-3-phosphate dehydrogenase (GAPD) activity can be provided by two isozymes (GapA or GapC); GapC is a heteromeric protein that requires genes from two genomic loci. The contents of a BiGG knowledgebase can be converted to a map (right) to facilitate visual interpretation. Or a mathematical modeling formalism to develop and explore hypotheses, such as a stoichiometric matrix (bottom) that can be used to explore mass flow through the network. (Modified reproduction from Reed et al.33 with permission).