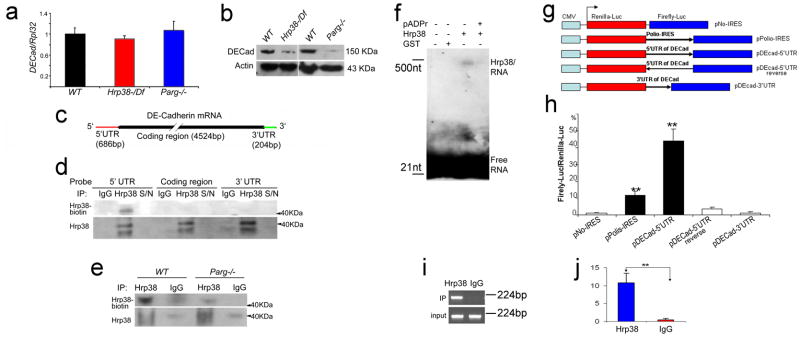
Figure 7. pADPr disrupts Hrp38 binding to 5′UTR of DE-cadherin conferring IRES activity.
(a) The mRNA expression level of DE-cadherin in the different genotypes at the wandering third-instar larvae stage. (b) The protein expression level of DE-cadherin in the different genotypes at the wandering third-instar larvae stage. (c) The structure of DE-cadherin transcript. The biotin-labeled probes were made from three regions (5′UTR, coding region and 3′UTR). (d) Hrp38 binding to 5′UTR of DE-cadherin in the ovary is shown by UV-crosslinking analysis. The ovarian lysate from the wild-type fly (y,w) crosslinked to the biotin-labeled RNA probes as indicated was IPed with rabbit anti-Hrp38 antibody or normal rabbit IgG as the control for IP. The supernatant (S/N), which was obtained after each IP, was run in the same blot to show the efficiency of RNase treatment. (e) Decreased amounts of Hrp38 protein binding to 5′UTR of DE-cadherin in the Parg mutant. In (d,e) Hrp38 protein binding to biotin-labeled RNA probe was detected with Streptavidin. The same blot was stripped and probed with anti-Hrp38 antibody to show IP efficiency. (f) Inhibition of Hrp38 binding to its target RNA motif by poly(ADP-ribose). The biotin-labeled G-rich RNA element (CAGGGCGCGCACUGUACGAG) within 5′UTR of DE-cadherin was incubated with the components as indicated. (g) Diagrams of the different constructs for dual luciferase assay. pNO-IRES (negtative control); pPolio-IRES (positive control); pDEcad 5′UTR: DE-cadherin 5′UTR cloned into the vector in the forward orientation. pDEcad 5′UTR-reverse (spacer control); pDEcad-3′UTR (element control). (h) The ratio of firefly-to-renilla luciferase activity after the transfection of the different constructs as indicated into Drosophila S2 cells. (i–j) The association of Hrp38 with the transcript from pDEcad 5′UTR-luciferase construct shown by regular RT-PCR (i) and qRT-PCR (j) after RNA-IP. RNA IP was done using anti-Hrp38 antibody or rabbit IgG as a control after the transfection of pDEcad-5′UTR construct into S2 cells. The error bars in (h,j) represents the standard deviation from three independent experiments. **P ≤ 0.01, analyzed by t-test.