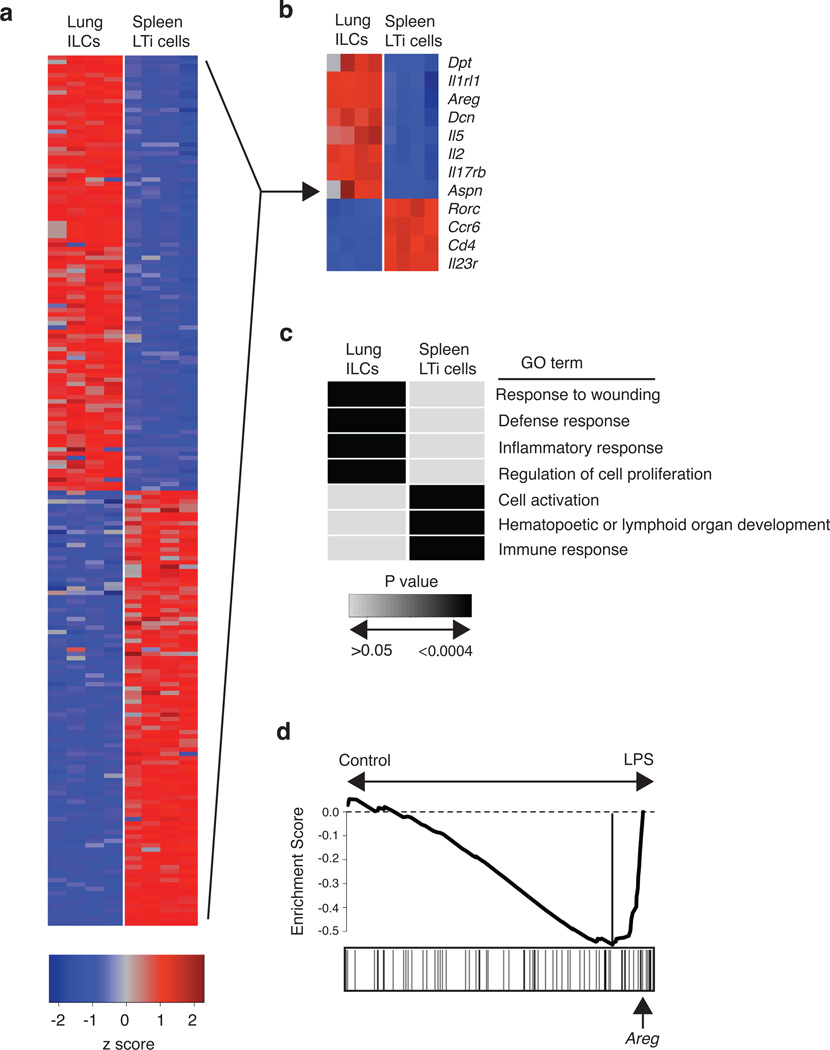
Figure 6. Global gene expression profiling of lung-resident ILCs reveals strong enrichment for genes regulating wound healing pathways.
(a-d) CD90+ CD25+ ILCs and CD90+ CD4+ LTi cells were FACS-sorted from the lung (ILCs) or spleen (LTi cells) of naïve C57BL/6 WT mice. mRNA was isolated, amplified, and hybridized to Affymetrix gene chips for microarray analysis. (a) Heat map representing gene expression profiles of the top 100 differentially expressed genes in lung ILCs versus spleen LTi cells. Red = high expression, blue = low expression. (b) Heat map of key genes highly expressed in lung ILCs or spleen LTi cells. Red = high expression, blue = low expression (c) Gene expression signatures of ILC and LTi cells (genes differentially expressed by two-fold or greater) were examined using Database for Annotation, Visualization and Integrated Discovery (DAVID) to identify enriched Gene Ontology terms describing biological processes. Shaded boxes represent significant enrichment (light gray P > 0.05, black P < 0.0004). (d) Gene Set Enrichment Analysis comparing the lung ILC gene expression signature with a previously published data set examining the effects of LPS-induced acute lung injury. Analysis of the top transcripts in the LPS-treated group shows presence of amphiregulin (highlighted by arrow).