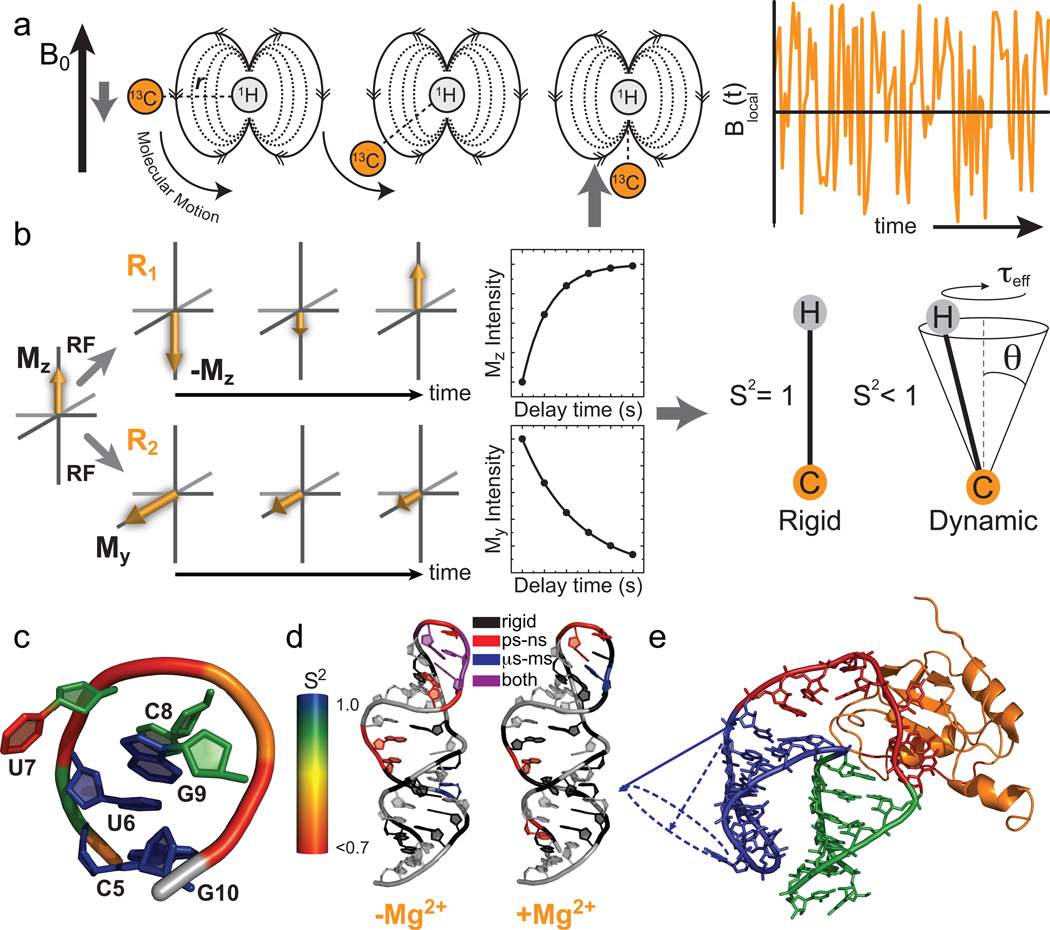
Figure 2.
Characterization of pico- to nanosecond motions using spin relaxation. (a) Reorientation of bond vectors leads to an oscillating local field at the nucleus of interest that influences relaxation. Shown is an example involving a 13C-1H dipolar interaction. (b) Model free analysis of longitudinal (R1) and transverse (R2) spin relaxation data yield an order parameter (S2) describing the amplitude of motion and a constant (τeff) describing its timescale. (c–e) Example applications of spin relaxation in studies of RNA dynamics, (c) Site-specific UUCG tetraloop dynamics. Note, the sugar and nucleobase order parameters17,32 were determined at 298 K while the phosphate backbone order parameters18 were determined at 310 K. (d) Re-distribution of motional modes in the catalytic domain 5 RNA of a group II intron upon addition of Mg2+ (Adapted from ref. 33 with permission from Elsevier). (e) RNA inter-helical motions persist when bound to the human U1A protein (Reprinted with permission from ref. 39. Copyright 2007 American Chemical Society).