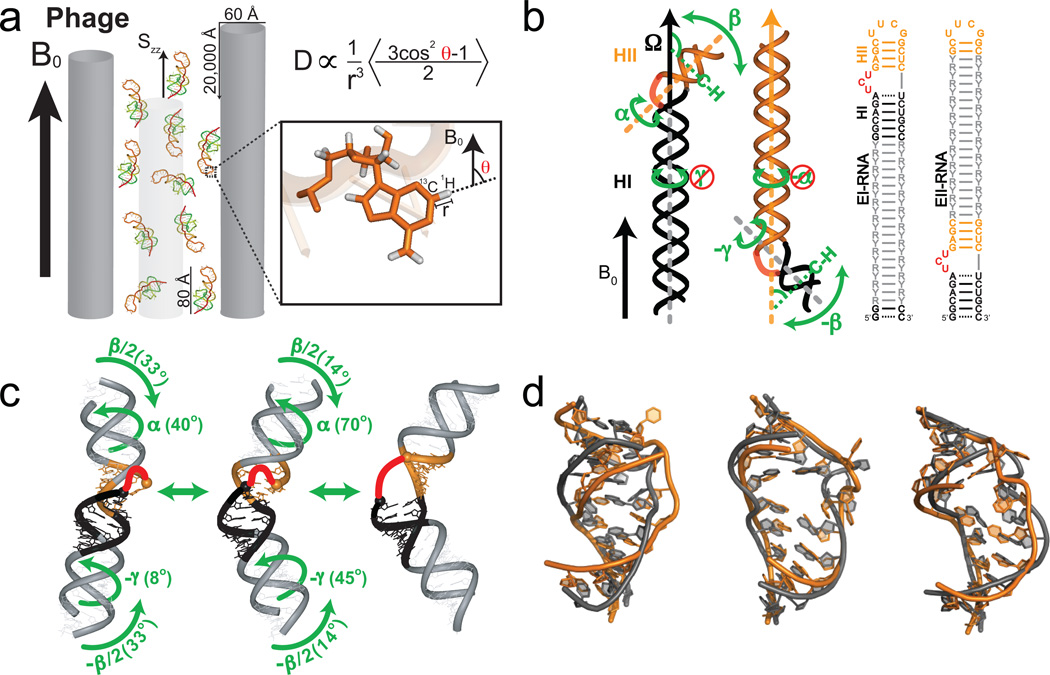
Figure 3.
Characterizing motions over sub-millisecond timescales using residual dipolar couplings. (a) Partial alignment of RNA using Pf1 phage shown as gray rods (left) (Adapted from ref. 43). (b) Domain-elongation for decoupling internal and overall motions allows measurement of bond vector dynamics relative to the elongated helix. An isotopic labeling strategy is used to render elongation residues (R-Y) NMR invisible (adapted from ref. 52). (c) Spatially correlated inter-helical motions observed using RDCs involving correlated changes in the inter-helical twist (α and γ) and bend (β) angles (adapted from ref. 52). (d) Combining domain-elongation RDCs and MD simulations in the construction of atomic-resolution dynamic ensembles of TAR reveals conformations (in gray) very similar to those observed in ligand bound states (in orange) (Adapted from ref. 55 by permission of Oxford University Press).