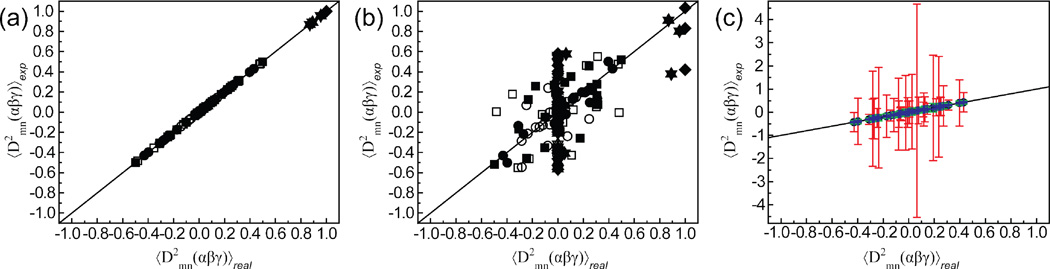
Figure 3.
Comparison of “real” Wigner rotation elements with “experimental” values computed using Equations 4, 7, and 9 based on (a) five and (b) four linearly independent alignment conditions. Results are shown for three motional models; continuous rotation of a domain by ± 25° about an arbitrary axis (square); collective motions of two helical domains in a 65 ns molecular dynamics simulation of TAR RNA25,34(circle); and ± 30° anti-correlated bending motions for domains I–II (triangle up), II–III (triangle down), and I–III (diamond). (c) The effects of RDC error and structural noise on Wigner elements determined from the MD trajectory for three levels of uncertainty in the alignment tensors, (blue error bars) ≤ 5° uncertainty in orientation and ≤ 5 % uncertainty in magnitude, (green error bars) 5°–10° uncertainty in orientation and 5–10 % uncertainty in magnitude, (red error bars) 10°–15° uncertainty in orientation and 10–15 % uncertainty in magnitude. Error bars represent the RMSD calculated over 100 random iterations.