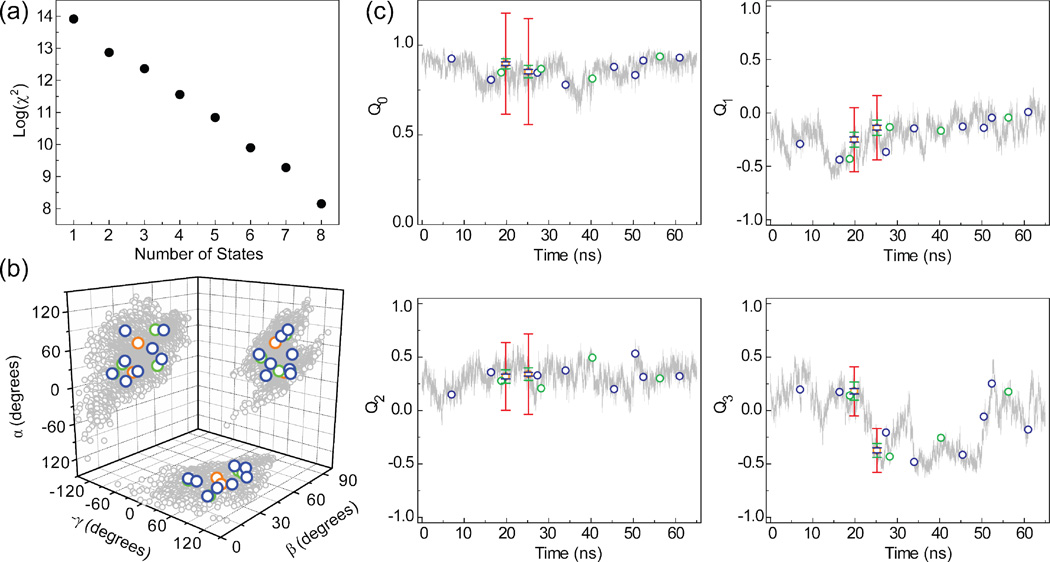
Figure 5.
n-state ensemble fit to computed for a motional model involving collective motions of two helical domains in a 65 ns molecular dynamics simulation of TAR RNA25,34. (a) χ2 value as a function of the number of states in the ensemble. (b) Comparison of the MD trajectory (shown in gray) and 2 (orange circles), 4 (green circles) and 8 (blue circles) state fits using an Euler angle representation described previously43. (c) Comparison of the fitted states to 64,998 state MD trajectory. Color coding is as in (b). Each state defined as an inter-helical domain orientation is represented using a unit quaternion; a 4-dimensional complex number that represents an orientation as a single axis rotation from the z-axis. The relationship between the quaternions and the Euler angles is given in Equation 14. Shown are the values for the four quaternion components; Q0, Q1, Q2, and Q3. Q0 encodes the rotational amplitude and Q1, Q2, and Q3 encode the x, y and z components of the rotation axis, respectively. Error bars are shown for the 2 state fit and represent the RMSD calculated over 100 random iterations. Color coding is as Figure 3. The order of the two states was chosen to minimize the overall RMSD compared to the zero error ensemble.