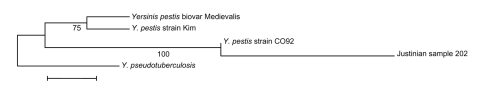
Figure.
Unrooted tree showing the phylogenetic relationships between the sequence obtained from the YP3 spacer from Justinian sample 202 and that from the genomes of Yersinia pestis strain CO92 (GenBank accession no. AJ414159), Y. pestis biovar Medievalis (AE017139), Y. pestis strain Kim (AE013993), and Y. pseudotuberculosis (BX936398). DNA sequences were aligned by using the ClustalW software, version 1.81 (2). Deletions were considered single events. A distance matrix was constructed by using the Kimura-2 parameter, and the phylogenetic tree was inferred by using the neighbor-joining method in the Mega2 software package. The scale bar represents a 0.5% nucleotide sequence divergence. Bootstrap values are indicated at the nodes (2).