Figure 1. Schematic diagram of the primer design process.
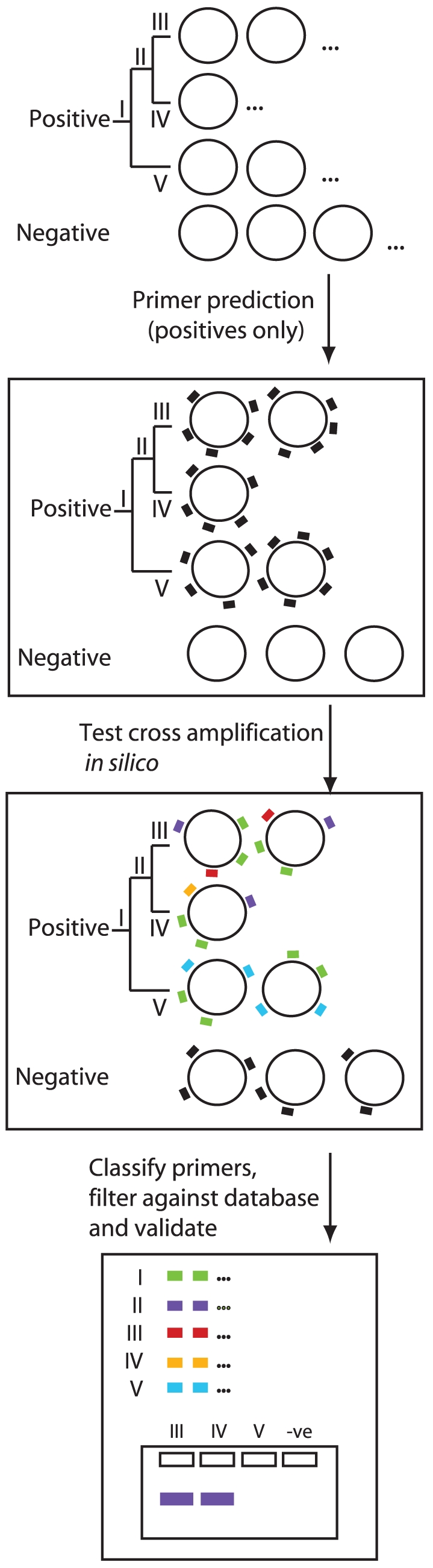
A training set of whole (complete or draft) genome sequences is divided into positive and negative sequence groups. Members of the positive sequence group are placed into classes as appropriate (here as classes I–V on the basis of a nested hierarchical relationship). Primer sets are designed to all positive sequences in bulk (>1000 primer sets, black markers), and tested for cross-hybridisation in silico. Primer sets that amplify only members of a prescribed class (indicated by coloured markers, one for each class; black markers indicate non-specific primers) but do not amplify negative examples are retained as being potentially diagnostic of that class. Predicted discriminatory primers are validated against bacterial isolates that were not part of the training set. An expected mock PCR result is indicated for primers specific to group II against individual samples belonging to classes II, IV and V. A detailed description of the method is given in Supplementary Methods.
