Abstract
The sequence of microbial genomes made all potential antigens of each pathogen available for vaccine development. This increased by orders of magnitude potential vaccine targets in bacteria, parasites, and large viruses and revealed virtually all their CD4+ and CD8+ T cell epitopes. The genomic information was first used for the development of a vaccine against serogroup B meningococcus, and it is now being used for several other bacterial vaccines. In this review, we will first summarize the impact that genome sequencing has had on vaccine development, and then we will analyze how the genomic information can help further our understanding of immunity to infection or vaccination and lead to the design of better vaccines by diving into the world of T cell immunity.
From Pasteur to Reverse Vaccinology
Vaccination is a medical practice of ancient origin that possibly started somewhere in Asia using materials from smallpox lesions to transmit a mild infection and thereby protect against more serious disease (Fenner et al., 1988). The practice was formally introduced into Western medicine in 1796 by Edward Jenner, who used infected materials isolated from cows (vacca in Latin) to immunize against smallpox and introduced the terminology “vaccine” (Jenner, 1801, 1959). A century later, when it was discovered that infections are caused by microbes, Louis Pasteur started the rational development of vaccines and established the basic rules of vaccinology (Pasteur, 1880). Pasteur proposed that in order to make a vaccine, one should “isolate, inactivate and inject the microorganism” (Rappuoli, 2004) that causes the disease. Pasteur’s rules were followed for a century by vaccine developers, from Jonas Salk, who developed a vaccine containing a poliovirus that had been killed by formaldehyde treatment, to Albert Sabin (Levine et al., 2009), who used a poliovirus that had been attenuated by serial passage in vitro. Hilleman developed vaccines against measles, mumps, and rubella by attenuating the viruses causing the diseases (Offit, 2007). Others, such as Ramon and Glenny, isolated essential components from bacterial or viral cultures, inactivated them, and paved the way for the development of vaccines against diphtheria and tetanus (Glenny and Hopkins, 1923, Ramon 1924), Neisseria meningitidis, Streptococcus pneumoniae, Haemophilus influenzae, and so on. In the case of hepatitis B, it was found that the causative virus could not be cultured in vitro. As a result, the vaccine was initially developed by inactivating viral antigen present in the plasma of chronically infected people (Buynak et al., 1976). The vaccines developed using Pasteur’s rules became powerful tools in the history of medicine and, in less than a century, led to the elimination of some of the most devastating infectious diseases globally.
At the end of the 20th century, most of the vaccines that could be developed by these traditional technologies had been developed, and new technologies were required to conquer the remaining pathogens. Remarkable progress was made during this period by the introduction of new technologies such as recombinant DNA and chemical conjugation of proteins to polysaccharides, as well as advances in the use of novel adjuvants. Additionally, a powerful tool came from the ability to access the genomes of microorganisms, a new technology that become available in 1995 when Craig Venter published the genome of the first free living organism (Fleischmann et al., 1995). This technological revolution allowed for the first time the capacity to move beyond the rules of Pasteur, using the computer to rationally design vaccines starting with information present in the genome, without the need to grow the specific microorganisms. This new approach was denominated “reverse vaccinology” (Rappuoli, 2000).
The first pathogen addressed by the reverse vaccinology approach was Meningococcus B (MenB), a pathogen that causes 50% of the meningococcal meningitis worldwide. This bacterium had been refractory to vaccine development because its capsular polysaccharide is identical to a human self-antigen, whereas the bacterial surface proteins are extremely variable. By mid 1990s, when the many attempts to develop a vaccine using the traditional technologies had failed and the hope to develop a vaccine was very low, we and others at TIGR began a project to sequence the MenB genome and to use the genomic information to develop a vaccine that had been refractory to development. Gene sequences were analyzed, and over 600 potential antigens were tested for antigenicity. Candidate sequences were expressed in Escherichia coli, and sera from immunized mice was obtained against each of them. Analysis of the sera revealed more than 90 previously unknown surface located proteins and that 29 of them were able to induce antibodies able to kill the bacteria in vitro in the presence of complement. The power of the technology became evident by noting that, up to that moment, only 12 surface antigens of meningococcus had been described in the literature, and of these, only 4 or 5 had bactericidal activity (Tettelin et al., 2000; Pizza et al., 2000). In subsequent years, the antigens discovered by this approach inducing the best and broadest bactericidal activity were selected and inserted into prototype vaccines that were able to induce protective immunity against most of the MenB strains in mice (Giuliani et al., 2006). After successful preclinical studies, the MenB vaccine entered the long path of vaccine development that included testing safety and immunogenicity in adult volunteers, initial testing in infants, and finally, a large scale, phase III clinical trial that is the basis for a European license application.
In the meantime, reverse vaccinology has been applied to many other bacterial pathogens. In the case of group B streptococcus, the analysis of eight genomes led to the expression of 312 surface proteins and the development of a vaccine composed of four proteins able to protect against all serotypes (Maione et al., 2005). A vaccine made by combination of a few bacterial proteins has also been developed for group A Streptococcus. In this case, because the infection is known to induce antibodies that cross-react with human antigens, the reverse vaccinology approach was essential to make sure that the selected antigens did not have homology to proteins encoded by the human genome. A genome-based approach is also being used to develop protein-based vaccines against emerging pathogens, such as antibiotic-resistant Staphylococcus aureus and Streptococcus pneumoniae. A vaccine which uses this approach against Chlamydia has also been described (Thorpe et al., 2007).
More recently, the power of the genome has been complemented by the ability to interrogate the entire antigenic repertoire. This has been done by using libraries of expressed antigens and screening for the immunogenicity of the proteins (referred to as the “antigenome”) during infection (Giefing et al., 2008), or by testing directly for the presence and amount of antigens on the surface of bacteria. This latter technology takes advantage of the power of mass spectrometry to identify and quantify those antigens that are present on the bacterial surface (Rodríguez-Ortega et al., 2006). Surface proteins of gram-positive bacteria are first partially digested by treatment with proteases, and then the resulting peptides are loaded on a mass spectrometer that provides information on the identity and quantity of surface exposed proteins. In the case of the gram-negative bacteria, antigens on the surface are identified by mass spectrometry of membrane fragments free of cytoplasmic material that are released by gram-negative bacteria, which have been genetically modified to weaken the outer membrane stability (Berlanda Scorza et al., 2008).
In addition to the discovery of many previously unknown antigens which have led to successful vaccine development in several instances, reverse vaccinology has made possible studies on antigen function, leading to an understanding of the biology of the pathogen. The two most notable examples are the discovery of pili in gram-positive pathogens and the discovery of factor H binding protein in meningococcus. In the first case, while pili had been known for decades to be an essential component for the pathogenesis of gram-negative bacteria, they were not known to be present in gram-positive pathogens such as group B and group A streptococcus and pneumococcus. However, following screening for protective antigens by reverse vaccinology, it was found that a protective antigen of group B streptococcus was a component of a high molecular weight pilus that is essential to mediate adhesion during colonization (Lauer et al., 2005). Following this initial observation, a pilus was also found in group A streptococcus and pneumococcus, revealing a unique mechanism of pathogenesis for these three important human pathogens. In the case of meningococcus, following the publication of the protective antigen GNA1870, two independent laboratories found that this antigen binds the human complement regulator factor H. This discovery led to an understanding that meningococcus can grow in human blood by downregulating the alternative pathway of complement activation. However, because the same protein was unable to bind Factor H (Madico et al., 2006; Schneider et al., 2009) from animal species such as mice and rats, the discovery also allowed the understanding of the species specificity of meningococcus and that failure to develop an animal model for meningococcus was because of the fact that the bacterium is unable to grow in the blood of mice and rats. Transgenic animals expressing human factor H may likely to be the solution for a meningococcus animal model.
In conclusion, reverse vaccinology uses the entire protein repertoire of each pathogen to select the best candidate vaccine antigens. This allows the development of vaccines that were previously difficult or impossible to make and can lead to the discovery of unique antigens that may improve existing vaccines. Processes affecting vaccine development during reverse vaccinology are reported in Figure 1. The main differences between conventional and reverse vaccinology are summarized in Table 1.
Figure 1. Schematic Diagram Summarizing the Pathway of Vaccine Development Starting from Reverse Vaccinology.
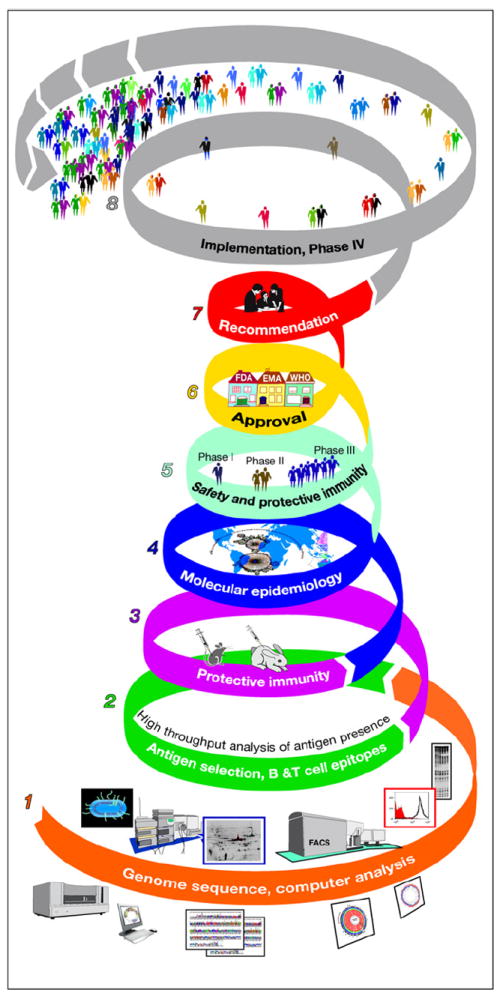
(1) First, computer analysis of the whole genome identifies the genes coding for predicted antigens and eliminates antigens with homologies to human proteins. (2) Then the identified antigens are screened for expression by the pathogen and for immunogenicity during infection. (3) The selected antigens are then used to immunize animals and test whether immunization induces a protective response. (4) Protective antigens are then tested for their presence and conservation in a collection of strains representative of the species (molecular epidemiology). (5) Finally, selected antigens are manufactured in large scale for clinical trials, and candidate vaccines are tested for safety and protective immunity in humans using established correlates of protection or efficacy studies. (6) Scientific, clinical, and technical information is then analyzed and approved by regulatory agencies, such as the Food and Drug Administration (FDA) or the European Medicinal Agency (EMA). (7) Policy-making bodies, such as the ACIP and equivalent bodies from other nations, make the recommendation on how the vaccine should be used. (8) The approved vaccine is then commercialized and used in large scale. At this point, phase IV clinical studies confirm safety.
Table 1.
Comparison between Traditional and Reverse Vaccinology
| Traditional Vaccinology | Reverse Vaccinology | |
|---|---|---|
| Antigens available | 10-25 identified by biochemical or genetic tools. | Virtually all antigens encoded by the genome are available. |
| Property of antigens | The most abundant antigens, the most immunogenic during disease, only from cultivable microorganisms. | All antigens are available, even if not highly immunogenic during disease. Antigens from noncultivable microorganisms can be identified. |
| Immunology of the antigens | Highly immunogenic antigens, often variable in sequence, because of immune selective pressure. Some may contain domains mimicking self-antigens and may induce autoimmunity. | The most conserved protective antigens can be identified. Usually these are not the most immunogenic during infection. The novel antigens are screened against the human genome, and antigens with homology to self-antigens are removed upfront |
| Polysaccharide antigens | A major target of traditional bacterial vaccines. | Cannot be identified by reverse vaccinology; however, operons coding for the biosynthesis of polysaccharides can be identified. This can lead to discovery of novel carbohydrate antigens. |
| T cell epitopes | Known epitopes limited to the known antigens. | Virtually every single T cell epitope is available. Screening of the total T cell immunity can be done by overlapping peptides. |
Measuring Cellular Immunity as a Tool to Understand and Design Responses to Pathogens and Vaccines
So far we have described how genome sequences allowed the discovery of previously unknown antigens and how this resulted in the development of vaccines against pathogens for which vaccines were not yet available. These vaccines work mainly by inducing serum antibodies, a necessary and often sufficient component of vaccine efficacy that can inactivate pathogens directly or by cooperation with complement or immune cells. However, it is well known that antibodies cannot be efficiently induced in the absence of CD4+ T cells that play a key role in B cell expansion and differentiation, class switching, and affinity maturation of the responses. In addition, elicitation of direct CD4+ T cell effector activity is important in vaccination strategies against tuberculosis, malaria, and parasitic diseases, while CD8+T cellular responses are a key element of the immune reactivity elicited by several vaccines, such as most attenuated vaccines.
In light of the important role of cellular immunity in vaccine efficacy and design, it seems logical and necessary, at least in certain cases, to also incorporate a cellular immunity dimension into reverse vaccinology. In this field, the genomic information, which contains the sequences of all known and unknown protein antigens of each pathogen, enabled the prediction of the entire repertoire of CD4+ T cell and CD8+ T cell epitopes by bioinformatic approaches and enabled their analysis by the synthesis of overlapping peptides. However, accurately measuring cellular immunity against complex pathogens is technically challenging and, until recently, has not been readily feasible. In this section, we describe some key examples of how measuring cellular immunity can provide insights into immunity and host-pathogen interactions and may inform the design of vaccines inducing optimal T cell responses.
In the last 10–15 years, the introduction of and optimization of technologies, such as tetrameric staining reagents, ELISPOT, and Intracellular Cytokine Staining (ICCS), have allowed for the exact measurement of cellular immune responses. This, in turn, has provided insights into the evolution of responses and enabled studies regarding pathogenesis, immunity, and correlates of protection. Monitoring cellular immunity is of particular importance in cases where CD4+T cell or CD8+ T cell responses are thought to be of relevance for protection and clearance of infection, as well as the establishment of chronic pathology (in the case of aberrant T cell responses). Examples include viral and bacterial infections (e.g., HIV, HCV, TB), certain parasitic diseases (malaria and several others), as well as autoimmunity and cancer. Although a comprehensive review of this topic is beyond the scope of this paper, we would like to highlight some specific examples.
It is widely acknowledged that cellular immunity in general, and CD8+T cell responses in particular, play an important role in the control of HIV (and SIV) replication and in establishing the viral set point ultimately determinant for clinical outcomes. Accordingly, studies analyzing epitope specific responses have revealed complex interactions between HIV and SIV and their hosts. For example, identification of the specific SIV epitopes within Tat and Gag which are recognized in the context of the MHC Mamu A*01 expressed by SIV infected macaques shed light onto the shift of responses from one antigen to another, the predictable mutation of dominant epitopes, and viral escape (Allen et al., 2000, 2001). In humans, several studies enabled by precise identification of epitopes recognized in the context of certain HLA types have highlighted how CD8+ T cell responses to different HIV proteins have discordant associations with viral load (Berger et al., 2010; Bhattacharya et al., 2007; Kiepiela et al., 2007; Zuñiga et al., 2006) and how certain mutations originate and are maintained within a given host but can actually revert upon transmission to a host lacking expression of the HLA type in question (Friedrich et al., 2004; Kawashima et al., 2009; Leslie et al., 2004). Based on the predicted and observed breadth of epitope recognition associated with different HLAs, it was recently suggested that thymic selection of the T cell repertoire might reflect the association of certain HLA alleles (John et al., 2010) with control of HIV infection (Kosmrlj et al., 2010).
Many studies in HIV and other systems have utilized pools of overlapping peptides to accurately characterize T cell responses, demonstrating that is not necessary to identify the actual epitope to measure responses (Betts et al., 2001; Maecker et al., 2001; Provenzano et al., 2007; Tobery et al., 2001). However, the large number of peptides required by this type of approach precludes its use to accurately characterize T cell responses in the case of large pathogens encoding hundreds or thousands of different ORFs.
Several studies have compared immune responses associated with positive clinical outcomes, pathogen clearance, and/or disease resolution and, in particular, examined if certain epitopes are differentially recognized. In general, differential recognition has not been observed. However, examining immune responses at the epitope level revealed differences in the quality of the T cell responses associated with different clinical outcomes (Appay et al., 2008; Harari et al., 2006, 2007). Examples of this line of experimentation are provided by studies that emphasized a correlation between the multispecificity of T cells and more favorable clinical outcomes in the case of HIV infection (Betts et al., 2006; Duvall et al., 2008; Zimmerli et al., 2005).
Tetrameric staining reagents allow the precise enumeration and characterization of T cells. Furthermore, as their use is dependent on TCR expression and not any specific functional activity, these reagents have been invaluable in instances where the T cells are rendered unresponsive as, for example, in the case of the regulation of the programmed death receptor 1(PD1) and its ligand, PD1L system associated with certain chronic infections (Barber et al., 2006; Day et al., 2006; Sester et al., 2008). Vaccine trials in humans, and previous trials in nonhuman primates (Manuel et al., 2009; Martins et al., 2010; Robb, 2008; Santra et al., 2010; Wilson et al., 2009), have monitored cellular responses using tetramers, isolated epitopes, and/or peptide pools.
Following epitope-specific responses is particularly informative for monitoring possible epitope loss or escape associated, for example, with vaccination against highly variable viruses or tumor epitopes. Monitoring different epitope-specific responses is also key to appreciate the degree to which epitope spreading, defined as the sequential appearance of new epitope-specific responses as a function of the evolution and maturation of cellular responses (Lehmann et al., 1993), is occurring, both molecularly and intramolecularly.
Reverse Vaccinology and Cellular Immunity
As discussed above, reverse vaccinology relies on the combined use of immunological and genomic information to identify relevant protein antigens for diagnostic or vaccine purposes. In this context, the identification of the epitopes recognized by CD4+ T cell or CD8+ T cells can be utilized in “reverse” as a tool to identify new antigens (Buus, 1999; De Groot et al., 2009b; Doolan et al., 2003; Lauemøller et al., 2000).
Pools of peptides can be designed to include peptides predicted to bind specific common HLA types. Alternatively, pools may be composed of peptides predicted to bind multiple alleles within an HLA supertype and thereby provide coverage representative of all populations without ethnic bias (Doytchinova and Flower, 2005; Doytchinova et al., 2004; Hertz and Yanover, 2007; Kangueane et al., 2005; Lund et al., 2004; Nielsen et al., 2010; Ou et al., 1998; Reche and Reinherz, 2004; Sette and Sidney, 1998, 1999; Sidney et al., 2008; Southwood et al., 1998; Tong et al., 2007). At the same time, pools may be formulated to include peptides representing a single putative antigen predicted on the basis of genomic sequence information. Pools can then be screened for immune reactivity with PBMC of exposed or vaccinated donors. If an antigen is immunodominant and is frequently recognized in the process of natural infection (or vaccination or exposure, depending on the study design), this strategy would lead to a “hit” even if the accuracy of the predictions is as low as 10%. As discussed below, consensus from several independent studies reveals that the actual success rate is indeed much higher.
An important issue to be addressed in the context of epitope prediction studies is the extremely large degree of polymorphism associated with HLA molecules. Indeed, thousands of different allelic molecules exist, each associated with a distinct peptide-binding specificity. Only a minor sample has been studied to date. Furthermore, even if predictions for thousands of different variants were available, synthesis and testing of thousands of different peptide sets would be clearly unpractical. However, two different approaches have been devised to overcome this difficulty. First, it has been noted that while thousands of alleles exist, a limited number (about 10 or 15, depending on the class and locus considered) allow coverage of the majority of the population (see, e.g., Sette and Sidney, 1998, 1999; Doolan et al., 2008; Southwood et al., 1998). Second, it has also been noted that although different alleles are each associated with a distinct binding specificity, most HLA alleles can be categorized into HLA “supertypes” associated with largely overlapping peptide specificity (HLA supermotifs; see, e.g., Doytchinova and Flower, 2005; Doytchinova et al., 2004; Hertz and Yanover, 2007; Kangueane et al., 2005; Lund et al., 2004; Nielsen et al., 2010; Ou et al., 1998; Reche and Reinherz, 2004; Sette and Sidney, 1998, 1999; Sidney et al., 2008; Southwood et al., 1998; Tong et al., 2007). Targeting these groups of alleles and associated motifs allows coverage of nearly of 90% of the general population with only a few peptide motifs. Furthermore, although the frequency of different alleles can vary dramatically in different ethnicities, the frequency of the supertype is relatively constant across different populations, thus avoiding the potential danger of ethnically biased population coverage.
By designing peptide pools to focus on HLA supertypes or the most common HLA types, there is no need for HLA typing PBMC donors as an inclusion criterion for these types of studies. Because relatively large pools of peptides can be easily assayed, the method allows one to quickly identify “hot spots” or immunodominant protein antigens. Accordingly, efforts can be directed at tackling larger genomes, previously unapproachable to systematic study because of their size and because of resource limitations.
In parallel with epitope and antigen mapping studies, it is important to conduct additional studies further validating the role of the cellular responses in immunity and protection. Studies such as depletion of CD4+ or CD8+ T cells, adoptive transfer experiments, or immunization with isolated epitopes can help assessing whether those responses are useful in preventing or attenuating disease (Yauch et al., 2009; Moutaftsi et al., 2009).
Knowing which epitopes are presented by infected cells, as opposed to cross-presented, may be critical to determine vaccine design. This has been addressed in the VACV system by examining the kinetics of antigen presentation in conditions favoring cross-presentation versus recognition of infected target cells (Gasteiger et al., 2007). Additional studies have analyzed the protective capacity of different VACV epitopes and found that the best correlates of protective capacity were high immunogenicity and capacity of being presented by infected cells (Moutaftsi et al., 2009).
Validations of the Epitope Prediction Approach
As a result of the considerable effort that has been devoted to developing epitope prediction technologies, most common HLA class I and class II alleles are covered by predictive algorithms publicly available at several sites, including the IEDB Analysis Resource (Zhang et al., 2008) (http://tools.immuneepitope.org). Several large-scale evaluations of available methods for predicting peptide binding to class I and class II MHC have been performed (Lin et al., 2008a, 2008b; Peters et al., 2006; Wang et al., 2008). For MHC class I, binding predictions generally perform extremely well, approaching error rates similar to what is observed between repeat experiments. Further algorithmic improvements for these predictions will, therefore, have little practical relevance. For class II molecules, binding predictions performed well, but there was still a substantial performance gap compared to the class I predictions and will, therefore, require further improvements.
Although the reason for the discrepancy in predictive accuracy between class I and class II molecules is not completely understood, it is widely believed that it might reflect multiple factors, all related to basic differences in the way the two classes bind their peptide ligands. First, the binding groove of class II molecules is open at both ends, allowing peptides of varying size to bind. Also, the peptide regions flanking the core region engaged by the MHC peptide-binding groove can influence binding in subtle and not easily predictable fashion. Furthermore, because the size of the typical class II peptide ligand is about 15 residues in size, or even longer, more than one binding register (each register corresponding to a 9-mer binding core) can be present in one peptide, further complicating the task of predicting binding affinity.
One promising strategy is the generation of consensus approaches that combine multiple prediction methods. Several studies have shown how consensus approaches can outperform individual predictors (Karpenko et al., 2008; Mallios, 2003; Moutaftsi et al., 2006; Wang et al., 2008).
The value of predictive algorithms for identifying CD8+ T cell epitopes was tested experimentally in a study by Moutaftsi et al. (2006). In this study, it was demonstrated that a bioinformatic approach could efficiently identify the vast majority of CD8+ T cell epitopes derived from vaccinia virus Western Reserve (VACV-WR) in the H-2b mouse model. Specifically, bioinformatic predictions based on a consensus approach led to the identification of 49 different epitopes, and additional experiments demonstrated that the epitopes identified by the prediction method accounted for approximately 95% of the total response to VACV infection in vivo.
To compare the prediction and overlapping peptide approaches, Kotturi et al. (2007) screened both a set of peptides predicted to bind major histocompatibility complex class I and a complete set of overlapping 15-mer peptides spanning the entire LCMV proteome for their capacity to induce IFN-γ production by CD8+ T cells derived from LCMV-infected H-2b mice. In addition to nine previously described epitopes, 19 novel epitopes were identified. These epitopes accounted for the total CD8+ CD44hi T cell response. Interestingly, the top 1.2%, or 160, predicted peptides accounted for 89% of the total response. By contrast, the overlapping peptide approach required synthesis of 664 peptides (plus several hundreds more to define the minimal epitope recognized) and identified epitopes accounting for only about 65% of the response. The failure of the 15-mer peptides to identify more epitopes is most likely because of the fact that they are not the optimal size for class I presentation and, therefore, require further processing to generate a functional epitope.
In terms of applicability to class II responses, recent improvements in prediction algorithms have translated into increased reliability in predicting class II restricted responses. For example, consensus predictions by Larry Stern’s group identified class II restricted epitopes in the DR1 VACV system (Calvo-Calle et al., 2007). Similarly, in the case of MCMV, 15 I-Ab-restricted CD4+ T cell epitopes were identified using consensus predictions (Arens et al., 2008).
Some important caveats should be kept in mind when considering epitope predictions. First, most epitope prediction methods consider only primary sequence, and the impact of 3D structure is not taken into account. Whereas it is generally assumed that since T cells recognize short linear fragments, and thereby 3D accessibility is of limited impact on epitope prediction for T cells (unlike the case for B cell epitopes), this issue might be of relevance in particular cases, such as for peptides involved in disulfide bridges. An additional caveat is that epitopes encompassing posttranslation modifications are not predicted. Indeed, several T cell epitopes with posttranslation modifications have been described. In these cases, mass spectroscopy-based sequencing prediction methods have been demonstrated to be the most effective technique to map the specific epitopes recognized.
Taken together, these studies have shown that efficient tools and approaches for identifying immune epitopes are already available. With the rapidly growing knowledge base pertaining to epitopes, reverse vaccinology by making available the entire repertoire of CD4+ T cell and CD8+ T cell epitopes provides the unique opportunity to optimize vaccines by selecting the most appropriate sequences.
Cellular Immunology and Technological Advances: The Landmark Case of Vaccinia and Other Complex Viral Pathogens
Historically, preferential targets of genome wide analysis of cellular immunity have encompassed small to medium size viruses. This was dictated by various factors, which included limitations in terms of algorithm accuracy, cost of performing binding assays to weed out nonbinders resulting from inaccurate predictions, and also the sheer cost of the peptide synthesis. An additional, and formidable, limitation was the lack of accurate and quantitative assays to detect T cell responses. At the same time, for many pathogens, the lack of genomic information represented an insurmountable barrier.
However, in the course of the last ten years, this scenario has changed dramatically, and it continues to evolve, as discussed in the section below discussing future directions for cellular responses and reverse vaccinology. The accuracy and breadth of predictive algorithms has improved dramatically, as pointed out above, thus reducing the number of leads to be examined and, in many cases, even obviating the need to prescreen peptide libraries to eliminate nonbinders to the specific MHC(s) targeted. Peptide costs have also dropped dramatically, by almost an order of magnitude, enabling synthesis of tens of thousands of peptides at a reasonable, albeit still expensive, cost. To be able to synthesize and test thousands of peptides is key for reverse vaccinology for T cells because, even with the most accurate predictions, thousand of peptides are obviously still required to analyze pathogen genomes encoding hundreds or thousands of ORFs. Also, as discussed above, much more quantitative and sensitive techniques are available to detect T cell responses. Finally, the number of genomes for which complete sequencing information is available is increasing exponentially. As a result, it has become possible to more comprehensively approach larger and more complex pathogens. In a parallel technology application, synthesis of hundreds of microbial ORFs has been shown to be suitable for CD4+ T cell testing to define immunodominance and population prevalence at the ORF level. In this application, pathogen specific T cells are enriched and then interrogated with the pathogen proteome either as a library (Jing et al., 2005, 2007; Koelle et al., 2001, 2003) or as pseudolibrary made up of the predicted ORFeome (Jing et al., 2008).
Perhaps the paradigm that demonstrates the combined benefit of these technological advantages is represented by the case of vaccinia virus. It is well known that vaccinia virus was used to eradicate smallpox, but this remarkable feat was accomplished several decades ago, when the immunological tools available to the scientific community were less sophisticated than today. As a result, the mechanisms of vaccination were not completely understood and the targets of immunity were not fully elucidated. Vaccination with VACV elicits a vigorous humoral and cellular immune response, detectable for decades following VACV immunization (Amara et al., 2004; Crotty et al., 2003; Demkowicz et al., 1996; Hammarlund et al., 2003; el-Ad et al., 1990; Putz et al., 2005). The general consensus is that cellular, and possibly also humoral, immunity are key for clearance of poxviruses following natural infection or vaccination (Fang and Sigal, 2005; Panchanathan et al., 2008), whereas humoral immunity is the main mechanism conferring protection from subsequent exposure following vaccination.
A renewed concern over the potential use of smallpox as a bioterrorist weapon prompted a fresh reexamination of the targets and mechanisms associated with vaccinia-specific immune responses. From 2003 on, many laboratories have produced an impressive amount of work, yielding an unprecedented wealth of information pertaining to the targets of CD4+ and CD8+ T cell immunity in humans, nonhuman primates, and mice (Moutaftsi et al., 2010; Walsh et al., 2009). These studies revealed that certain antigens are more prevalently recognized by CD8+ T cell responses. The majority of these antigens tended to be expressed early in infection, although late antigens are sometimes also recognized (Jing et al., 2005; Moutaftsi et al., 2006; Oseroff et al., 2005). The antigens more prevalently recognized in the case of CD4+ T cell responses tend to be different from the ones recognized by CD8+ T cell responses (Moutaftsi et al., 2007). Interestingly, the antigens dominantly recognized by antibodies are similar to the ones recognized by CD4+ T cell responses (Jing et al., 2008; Sette et al., 2008). Abortive infection of DC by VACV could lead to a preference for early antigens by CD8+ T cell, whereas viral particles taken up by uninfected bystander DC (exogenous) lead to recognition of late antigens by CD4+ T cell (Chahroudi et al., 2006; Engelmayer et al., 1999; Jenne et al., 2000; Liu et al., 2005). Likewise, Sigal’s group recently showed that direct presentation is sufficient to generate VACV CD8+ T cell responses (Xu et al., 2010). One conclusion of the studies by Jing et al. (2008) was that the population of prevalent CD4+ T cell antigens also represented the most abundant virion proteins, as measured by weight percent or mole percent in purified virions.
Because similar patterns, in terms of the types of antigens recognized by the different response types (i.e., CD4+ T cell, antibody, CD8+ T cell), were observed in different animal species, this data provided an exemplary justification for the validation and use of animal models to measure and compare the performance of different vaccine candidates. A parallel set of studies led to the generation of a complete characterization of the VACV transcriptome, in terms of the kinetics of expression of the various encoded ORFs, a gene array analysis of the host response to infection, and proteomic analysis of VACV virions (Assarsson et al., 2008; Moutaftsi et al., 2010). Taken together, these data allowed for the first time, in a complex pathogen, precise correlation of humoral and cellular immunity with genomic, transcriptomic, and proteomic information. It was found that early (4 hr) mRNA correlates with recognition by CD8+ T cell responses, while levels of protein expression best correlates with recognition by antibodies and CD4+ T cell responses. It is tempting to correlate these results in light of the DRIP theory for CD8+ T cell recognition (Yewdell and Nicchitta, 2006), while speculating that the correlation between protein abundance and CD4+ T cell responses might be reflective of the fact that class II recognition is mostly associated with the exogenous antigen processing pathway. Furthermore, these results called into question the validity of a simple cognate antigen model for antibody and T cell responses, at least in the case of complex viruses (Sette et al., 2008). These data are of particular interest in the context of reverse vaccinology, as they demonstrate that different subsets of antigens might be relevant (and perhaps could be combined) in the context of different types of adaptive responses.
These issues are not only relevant in the context of VACV. Indeed, other demonstrations of how different types of adaptive responses might recognize different sets of antigens come from the study of herpes viruses. An important study (Gredmark-Russ et al., 2008) utilizing the technique of caged tetramer loading (Toebes et al., 2006), and a subsequent study utilizing epitope predictions (Freeman et al., 2010), characterized, for example, the targets of CD8 cellular immunity recognized in the case of the Murine gammaherpesvirus 68. Parallel studies probed other viruses of the herpes family and CD4+ T cell responses as well. These studies illustrate the complexity of responses involved and also how different kinetic classes are recognized, how different antigen/epitope specificities are associated with differential memory maturation (Arens et al., 2008), and how antigens expressed in different stages of infection can be associated with differential immunity (Freeman et al., 2010). Taken together, these studies emphasize the important differences in the quality of responses directed against different antigens in the case of complex viral pathogens.
In conclusion, it appears that combining epitope mapping and antigen identification studies with genetic and proteomic studies is required to pinpoint the kinetics, cell specificity, subcellular localization, and expression patterns of the various antigens and is required to provide a complete picture of the immune response to complex pathogens. This information, in turn, will be essential for rational vaccine design and to further our understanding of host pathogen interactions.
Examples of T Cell Vaccines
Genome sequences and the new immunological and bioinformatic tools described above expanded enormously the ability to analyze the cellular immune response to pathogens and vaccines; however, our ability to use this information to design better vaccines is still limited. Below are few examples of how T cell immunity can be used to design better vaccines.
The knowledge of promiscuous T cell epitopes was used to design a recombinant protein containing 9 different CD4+ T cell epitopes that was covalently linked to the hemophilus influenzae type b oligosaccharide to make a new conjugate vaccine that was capable of generating responses as good as the conjugate vaccine used contemporaneously for mass immunization (Falugi et al., 2001). An optimized nonnatural pan DR epitope (PADRE), which was shown to elicit better immune responses than several pathogen-derived promiscuous T helper cell epitopes, was incorporated into several vaccines in development (Alexander et al., 2000). Synthetic peptides or genes coming from the sequences of viral or bacterial pathogens have been used for vaccines against chronic infections such as hepatitis B, hepatitis C, and HIV (Moynihan and Howard, 2001; Klade et al., 2008; Graham et al., 2010). A T cell-based vaccine against HIV, delivered by an adenovirus vector, showed no protective effect against HIV, although it had been able to reduce the viral load in non human primates (Buchbinder et al., 2008). Finally peptide-based vaccines have shown some efficacy in clinical trials against cancer, and the efficacy appeared to correlate with induction of T cell-specific immunity (Bocchia et al., 2005; Kenter et al., 2009). These results are very promising because the ability to sequence the genome of cancer cells will increasingly provide new information on cancer-specific epitopes and increase our ability to design novel cancer vaccines.
Future Trends: Further Technology Development and Integration with Bioinformatics, Genomics, Proteomics, and Systems Biology
The outlook for reverse vaccinology is bright. A growing number of studies demonstrate that the technical and conceptual advances of recent years have enabled tackling large and complex pathogens, encoding thousands of different ORFs. Examples of these pathogens include bacterial and protozoan pathogens, such as mycobacteria, plasmodium, anthrax, and tularemia (Crompton et al., 2010; Doolan et al., 2003, 2008; Doolan et al., 2003; Ingram et al., 2010a, 2010b; Lewinsohn et al., 2007; McMurry et al., 2005, 2007; Moise et al., 2009).
Comprehensive meta-analyses of the immune epitope data published in the scientific literature have also provided valuable insights. For example, such an analysis in the context of both plasmodium and mycobacterium revealed that, in both cases, the described epitopes are mainly derived from a small fraction of the genome, despite the fact that these two pathogens encode thousands of ORFs (Blythe et al., 2007; Vaughan et al., 2009). For example, the MTB genome comprises approximately 4000 coding ORFs, from which 721 unique T cell epitopes are defined in the literature. However, the MTB epitopes are derived from relatively few ORFs. In total, as few as 30 ORFs account for 85% of the defined epitopes, and if the 93 known human CD4+ T cell epitopes are considered, as few as 10 ORFs account for 80% of the epitopes.
The explanation for the narrow breadth of reported responses against MTB might lie in historical considerations because soluble antigens and/or antigens identified early on were more thoroughly investigated. Alternatively, the narrow breadth of antigens could reflect a focus of the immune response on a few targets recognized as immunogenic (immunodominance), leading to the immune system being, in effect, blind or oblivious to 95% of the pathogen genome. However, recent studies in several complex pathogen systems revealed broad immune responses, directed against a relatively large fraction of the genome. Indeed, analysis using protein chip arrays, in the context of both plasmodium and mycobacteria, reveals a much broader spectrum of responses than originally suspected. Thus, these observations provide a strong rationale for performing truly unbiased analysis of cellular immunity in complex pathogens.
At the technical level, several different issues are coming to fruition. For example, the availability of tetramers is constantly expanding in several regards. First, the breadth of alleles for which multimeric staining reagents are commonly available is constantly expanding (Altman et al., 1996; Bakker and Schumacher, 2005; McHeyzer-Williams et al., 1996). Indeed, the most powerful way to demonstrate peptide-specific, MHC-restricted, T cell epitopes is obtained by HLA tetramers. MHC predictors are essential for the rational identification of both the optimal peptide and the restricting MHC. The ability to subsequently generate the appropriate HLA tetramers is essential for the proper implementation of this strategy (Leisner et al., 2008). Second, recent work in the class II system has shed new light on some of the technical challenges associated with tetramer production for Class II molecules (Landais et al., 2009). Furthermore, the use of tetramer reagents in tandem with epitope identification strategies represents a valuable approach to determining frequencies of naive T cells in unexposed individuals, in the context of class I (Kotturi et al., 2008) and also class II (Moon et al., 2007).
Another important issue is achieving sufficient coverage of MHC polymorphism. Decisive progress has been recently made in terms of the definition of motifs and development of bioinformatic tools, to allow predictions for a majority, or at least a large fraction, of all common HLA class I and class II molecules. Because of these recent advances, population coverage in excess of 90% at the class I A and B loci is now possible. Similarly, coverage of >80% of individuals at each class II locus, including the historically much underrepresented DQ and DP loci, is now achievable (P. Wang, J. Sidney, A.S., and B. Peters, unpublished data). Furthermore, in terms of characterizing the ever growing numbers of MHC alleles, new approaches to generate pan-specific predictors for both class I and II suggest that the end might be in sight for the long quest for a complete mapping of MHC specificities called for in the “Human MHC project” (Nielsen et al., 2007, 2008). Still, other areas need attention to expand the reverse vaccinology platform. For example, the definition of motifs, development of tools for predictions, and generation of reagents for tetramers for species other than mice and humans, such as ferrets, certain primates (Chinese macaques and cynomolgous monkeys), guinea pigs, and other species commonly utilized for evaluations of disease models and vaccine candidates, is essential.
An important development is the growing availability of bioinformatic resources that store and organize both immune reactivity data and pathogen data (Aurrecoechea et al., 2010; Greene et al., 2007; Snyder et al., 2007; Squires et al., 2008). This is particularly key as the amount of data escalate, both regarding immune recognition and genomic transcriptomic and proteomic information for various pathogens. The intersection of these two types of resources allows the development and testing of new hypotheses, such as those exemplified in the analysis of epitope information and influenza strain variation in the context of the recent swine origin flu pandemic (Greenbaum et al., 2009) and in the case of the design of promising new mosaic immunogens as potential HIV vaccines (Barouch et al., 2010; De Groot et al., 2005, 2009a; De Groot et al., 2005; Koita et al., 2006; Santra et al., 2010).
In conclusion, it is easy to predict that in the following years, an ever-growing widespread application of reverse vaccinology to infectious diseases in an integrated fashion. These advances will allow taking into account the heterogeneity of host responses in general and individual cellular responses in particular, but also, and quite importantly, pathogen heterogeneity. The lessons learned, and the technologies developed, will also be applicable to other diseases where strategies to either induce or control cellular immune responses are of potential clinical benefit, such as in autoimmunity and cancer.
Acknowledgments
We would like to thank M. Kotturi, J. Sidney, M. Moutaftsi, and B. Mothe for their comments and critical reading of the manuscript. The authors are also grateful to C. Mallia for editorial assistance and G. Corsi for the artwork. This work is supported by funding from NIH-NIAID contracts HHSN272200700048C, -900042C, -900044C (to A.S.).
References
- Alexander J, del Guercio MF, Maewal A, Qiao L, Fikes J, Chesnut RW, Paulson J, Bundle DR, DeFrees S, Sette A. Linear PADRE T helper epitope and carbohydrate B cell epitope conjugates induce specific high titer IgG antibody responses. J Immunol. 2000;164:1625–1633. doi: 10.4049/jimmunol.164.3.1625. [DOI] [PubMed] [Google Scholar]
- Allen TM, O’Connor DH, Jing P, Dzuris JL, Mothé BR, Vogel TU, Dunphy E, Liebl ME, Emerson C, Wilson N, et al. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature. 2000;407:386–390. doi: 10.1038/35030124. [DOI] [PubMed] [Google Scholar]
- Allen TM, Mothé BR, Sidney J, Jing P, Dzuris JL, Liebl ME, Vogel TU, O’Connor DH, Wang X, Wussow MC, et al. CD8(+) lymphocytes from simian immunodeficiency virus-infected rhesus macaques recognize 14 different epitopes bound by the major histocompatibility complex class I molecule mamu-A*01: implications for vaccine design and testing. J Virol. 2001;75:738–749. doi: 10.1128/JVI.75.2.738-749.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman JD, Moss PA, Goulder PJ, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ, Davis MM. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. doi: 10.1126/science.274.5284.94. [DOI] [PubMed] [Google Scholar]
- Amara RR, Nigam P, Sharma S, Liu J, Bostik V. Long-lived poxvirus immunity, robust CD4 help, and better persistence of CD4 than CD8 T cells. J Virol. 2004;78:3811–3816. doi: 10.1128/JVI.78.8.3811-3816.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appay V, van Lier RA, Sallusto F, Roederer M. Phenotype and function of human T lymphocyte subsets: consensus and issues. Cytometry A. 2008;73:975–983. doi: 10.1002/cyto.a.20643. [DOI] [PubMed] [Google Scholar]
- Arens R, Wang P, Sidney J, Loewendorf A, Sette A, Schoenberger SP, Peters B, Benedict CA. Cutting edge: murine cytomegalovirus induces a polyfunctional CD4 T cell response. J Immunol. 2008;180:6472–6476. doi: 10.4049/jimmunol.180.10.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assarsson E, Greenbaum JA, Sundström M, Schaffer L, Hammond JA, Pasquetto V, Oseroff C, Hendrickson RC, Lefkowitz EJ, Tscharke DC, et al. Kinetic analysis of a complete poxvirus transcriptome reveals an immediate-early class of genes. Proc Natl Acad Sci USA. 2008;105:2140–2145. doi: 10.1073/pnas.0711573105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurrecoechea C, Brestelli J, Brunk BP, Fischer S, Gajria B, Gao X, Gingle A, Grant G, Harb OS, Heiges M, et al. EuPathDB: a portal to eukaryotic pathogen databases. Nucleic Acids Res. 2010;38(Database issue):D415–D419. doi: 10.1093/nar/gkp941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker AH, Schumacher TN. MHC multimer technology: current status and future prospects. Curr Opin Immunol. 2005;17:428–433. doi: 10.1016/j.coi.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, Ahmed R. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439:682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- Barouch DH, O’Brien KL, Simmons NL, King SL, Abbink P, Maxfield LF, Sun YH, La Porte A, Riggs AM, Lynch DM, et al. Mosaic HIV-1 vaccines expand the breadth and depth of cellular immune responses in rhesus monkeys. Nat Med. 2010;16:319–323. doi: 10.1038/nm.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger CT, Carlson JM, Brumme CJ, Hartman KL, Brumme ZL, Henry LM, Rosato PC, Piechocka-Trocha A, Brockman MA, Harrigan PR, et al. Viral adaptation to immune selection pressure by HLA class I-restricted CTL responses targeting epitopes in HIV frameshift sequences. J Exp Med. 2010;207:61–75. doi: 10.1084/jem.20091808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlanda Scorza F, Doro F, Rodríguez-Ortega MJ, Stella M, Liberatori S, Taddei AR, Serino L, Gomes Moriel D, Nesta B, Fontana MR. Proteomics characterization of outer membrane vesicles from the extraintestinal pathogenic Escherichia coli DeltatolR IHE3034 mutant. Mol Cell Proteomics. 2008;7:473–485. doi: 10.1074/mcp.M700295-MCP200. [DOI] [PubMed] [Google Scholar]
- Betts MR, Ambrozak DR, Douek DC, Bonhoeffer S, Brenchley JM, Casazza JP, Koup RA, Picker LJ. Analysis of total human immunodeficiency virus (HIV)-specific CD4(+) and CD8(+) T-cell responses: relationship to viral load in untreated HIV infection. J Virol. 2001;75:11983–11991. doi: 10.1128/JVI.75.24.11983-11991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, Lederman MM, Benito JM, Goepfert PA, Connors M, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya T, Daniels M, Heckerman D, Foley B, Frahm N, Kadie C, Carlson J, Yusim K, McMahon B, Gaschen B, et al. Founder effects in the assessment of HIV polymorphisms and HLA allele associations. Science. 2007;315:1583–1586. doi: 10.1126/science.1131528. [DOI] [PubMed] [Google Scholar]
- Blythe MJ, Zhang Q, Vaughan K, de Castro R, Jr, Salimi N, Bui HH, Lewinsohn DM, Ernst JD, Peters B, Sette A. An analysis of the epitope knowledge related to Mycobacteria. Immunome Res. 2007;3:10. doi: 10.1186/1745-7580-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocchia M, Gentili S, Abruzzese E, Fanelli A, Iuliano F, Tabilio A, Amabile M, Forconi F, Gozzetti A, Raspadori D, et al. Effect of a p210 multipeptide vaccine associated with imatinib or interferon in patients with chronic myeloid leukaemia and persistent residual disease: a multicentre observational trial. Lancet. 2005;365:657–662. doi: 10.1016/S0140-6736(05)17945-8. [DOI] [PubMed] [Google Scholar]
- Buchbinder SP, Mehrotra DV, Duerr A, Fitzgerald DW, Mogg R, Li D, Gilbert PB, Lama JR, Marmor M, Del Rio C, et al. Step Study Protocol Team. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet. 2008;372:1881–1893. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buus S. Description and prediction of peptide-MHC binding: the ‘human MHC project’. Curr Opin Immunol. 1999;11:209–213. doi: 10.1016/s0952-7915(99)80035-1. [DOI] [PubMed] [Google Scholar]
- Buynak EB, Roehm RR, Tytell AA, Bertland AU, II, Lampson GP, Hilleman MR. Vaccine against human hepatitis B. JAMA. 1976;235:2832–2834. [PubMed] [Google Scholar]
- Calvo-Calle JM, Strug I, Nastke MD, Baker SP, Stern LJ. Human CD4+ T cell epitopes from vaccinia virus induced by vaccination or infection. PLoS Pathog. 2007;3:1511–1529. doi: 10.1371/journal.ppat.0030144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahroudi A, Garber DA, Reeves P, Liu L, Kalman D, Feinberg MB. Differences and similarities in viral life cycle progression and host cell physiology after infection of human dendritic cells with modified vaccinia virus Ankara and vaccinia virus. J Virol. 2006;80:8469–8481. doi: 10.1128/JVI.02749-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crompton PD, Kayala MA, Traore B, Kayentao K, Ongoiba A, Weiss GE, Molina DM, Burk CR, Waisberg M, Jasinskas A, et al. A prospective analysis of the Ab response to Plasmodium falciparum before and after a malaria season by protein microarray. Proc Natl Acad Sci USA. 2010;107:6958–6963. doi: 10.1073/pnas.1001323107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S, Felgner P, Davies H, Glidewell J, Villarreal L, Ahmed R. Cutting edge: long-term B cell memory in humans after smallpox vaccination. J Immunol. 2003;171:4969–4973. doi: 10.4049/jimmunol.171.10.4969. [DOI] [PubMed] [Google Scholar]
- Day CL, Kaufmann DE, Kiepiela P, Brown JA, Moodley ES, Reddy S, Mackey EW, Miller JD, Leslie AJ, DePierres C, et al. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature. 2006;443:350–354. doi: 10.1038/nature05115. [DOI] [PubMed] [Google Scholar]
- De Groot AS, Marcon L, Bishop EA, Rivera D, Kutzler M, Weiner DB, Martin W. HIV vaccine development by computer assisted design: the GAIA vaccine. Vaccine. 2005;23:2136–2148. doi: 10.1016/j.vaccine.2005.01.097. [DOI] [PubMed] [Google Scholar]
- De Groot AS, Ardito M, McClaine EM, Moise L, Martin WD. Immunoinformatic comparison of T-cell epitopes contained in novel swine-origin influenza A (H1N1) virus with epitopes in 2008-2009 conventional influenza vaccine. Vaccine. 2009a;27:5740–5747. doi: 10.1016/j.vaccine.2009.07.040. [DOI] [PubMed] [Google Scholar]
- De Groot ASMJ, Moise L, Martin B. Epitope-based Immunome-derived Vaccines: A Strategy for Improved Design and Safety. In: Falus A, editor. Clinical Applications of Immunomics. New York: Springer; 2009b. [Google Scholar]
- Demkowicz WE, Jr, Littaua RA, Wang J, Ennis FA. Human cytotoxic T-cell memory: long-lived responses to vaccinia virus. J Virol. 1996;70:2627–2631. doi: 10.1128/jvi.70.4.2627-2631.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolan DL, Southwood S, Freilich DA, Sidney J, Graber NL, Shatney L, Bebris L, Florens L, Dobano C, Witney AA, et al. Identification of Plasmodium falciparum antigens by antigenic analysis of genomic and proteomic data. Proc Natl Acad Sci USA. 2003;100:9952–9957. doi: 10.1073/pnas.1633254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolan DL, Mu Y, Unal B, Sundaresh S, Hirst S, Valdez C, Randall A, Molina D, Liang X, Freilich DA, et al. Profiling humoral immune responses to P. falciparum infection with protein microarrays. Proteomics. 2008;8:4680–4694. doi: 10.1002/pmic.200800194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doytchinova IA, Flower DR. In silico identification of supertypes for class II MHCs. J Immunol. 2005;174:7085–7095. doi: 10.4049/jimmunol.174.11.7085. [DOI] [PubMed] [Google Scholar]
- Doytchinova IA, Guan P, Flower DR. Identifiying human MHC supertypes using bioinformatic methods. J Immunol. 2004;172:4314–4323. doi: 10.4049/jimmunol.172.7.4314. [DOI] [PubMed] [Google Scholar]
- Duvall MG, Precopio ML, Ambrozak DA, Jaye A, McMichael AJ, Whittle HC, Roederer M, Rowland-Jones SL, Koup RA. Polyfunctional T cell responses are a hallmark of HIV-2 infection. Eur J Immunol. 2008;38:350–363. doi: 10.1002/eji.200737768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Ad B, Roth Y, Winder A, Tochner Z, Lublin-Tennenbaum T, Katz E, Schwartz T. The persistence of neutralizing antibodies after revaccination against smallpox. J Infect Dis. 1990;161:446–448. doi: 10.1093/infdis/161.3.446. [DOI] [PubMed] [Google Scholar]
- Engelmayer J, Larsson M, Subklewe M, Chahroudi A, Cox WI, Steinman RM, Bhardwaj N. Vaccinia virus inhibits the maturation of human dendritic cells: a novel mechanism of immune evasion. J Immunol. 1999;163:6762–6768. [PubMed] [Google Scholar]
- Falugi F, Petracca R, Mariani M, Luzzi E, Mancianti S, Carinci V, Melli ML, Finco O, Wack A, Di Tommaso A, et al. Rationally designed strings of promiscuous CD4(+) T cell epitopes provide help to Haemophilus influenzae type b oligosaccharide: a model for new conjugate vaccines. Eur J Immunol. 2001;31:3816–3824. doi: 10.1002/1521-4141(200112)31:12<3816::AID-IMMU3816>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Fang M, Sigal LJ. Antibodies and CD8+ T cells are complementary and essential for natural resistance to a highly lethal cytopathic virus. J Immunol. 2005;175:6829–6836. doi: 10.4049/jimmunol.175.10.6829. [DOI] [PubMed] [Google Scholar]
- Fenner F, Henderson DA, Arita L, Jezek Z, Ladnyi ID. Smallpox and its eradication. Geneva: World Health Organization; 1988. [Google Scholar]
- Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, Merrick JM, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- Freeman ML, Lanzer KG, Cookenham T, Peters B, Sidney J, Wu TT, Sun R, Woodland DL, Sette A, Blackman MA. Two kinetic patterns of epitope-specific CD8 T-cell responses following murine gammaherpesvirus 68 infection. J Virol. 2010;84:2881–2892. doi: 10.1128/JVI.02229-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich TC, Dodds EJ, Yant LJ, Vojnov L, Rudersdorf R, Cullen C, Evans DT, Desrosiers RC, Mothé BR, Sidney J, et al. Reversion of CTL escape-variant immunodeficiency viruses in vivo. Nat Med. 2004;10:275–281. doi: 10.1038/nm998. [DOI] [PubMed] [Google Scholar]
- Gasteiger G, Kastenmuller W, Ljapoci R, Sutter G, Drexler I. Cross-priming of cytotoxic T cells dictates antigen requisites for modified vaccinia virus Ankara vector vaccines. J Virol. 2007;81:11925–11936. doi: 10.1128/JVI.00903-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giefing C, Meinke AL, Hanner M, Henics T, Bui MD, Gelbmann D, Lundberg U, Senn BM, Schunn M, Habel A, et al. Discovery of a novel class of highly conserved vaccine antigens using genomic scale antigenic fingerprinting of pneumococcus with human antibodies. J Exp Med. 2008;205:117–131. doi: 10.1084/jem.20071168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliani MM, Adu-Bobie J, Comanducci M, Aricò B, Savino S, Santini L, Brunelli B, Bambini S, Biolchi A, Capecchi B, et al. A universal vaccine for serogroup B meningococcus. Proc Natl Acad Sci USA. 2006;103:10834–10839. doi: 10.1073/pnas.0603940103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenny AT, Hopkins BE. Diphtheria toxoid as an immunizing agent. Br J Exp Pathol. 1923;4:283–288. [Google Scholar]
- Graham BS, McElrath MJ, Keefer MC, Rybczyk K, Berger D, Weinhold KJ, Ottinger J, Ferarri G, Montefiori DC, Stablein D, et al. Immunization with Cocktail of HIV-Derived Peptides in Montanide ISA-51 Is Immunogenic, but Causes Sterile Abscesses and Unacceptable Reactogenicity. PLoS One. 2010;5:e11995. doi: 10.1371/journal.pone.0011995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gredmark-Russ S, Cheung EJ, Isaacson MK, Ploegh HL, Grotenbreg GM. The CD8 T-cell response against murine gammaherpesvirus 68 is directed toward a broad repertoire of epitopes from both early and late antigens. J Virol. 2008;82:12205–12212. doi: 10.1128/JVI.01463-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum JA, Kotturi MF, Kim Y, Oseroff C, Vaughan K, Salimi N, Vita R, Ponomarenko J, Scheuermann RH, Sette A, Peters B. Pre-existing immunity against swine-origin H1N1 influenza viruses in the general human population. Proc Natl Acad Sci USA. 2009;106:20365–20370. doi: 10.1073/pnas.0911580106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene JM, Collins F, Lefkowitz EJ, Roos D, Scheuermann RH, Sobral B, Stevens R, White O, Di Francesco V. National Institute of Allergy and Infectious Diseases bioinformatics resource centers: new assets for pathogen informatics. Infect Immun. 2007;75:3212–3219. doi: 10.1128/IAI.00105-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarlund E, Lewis MW, Hansen SG, Strelow LI, Nelson JA, Sexton GJ, Hanifin JM, Slifka MK. Duration of antiviral immunity after smallpox vaccination. Nat Med. 2003;9:1131–1137. doi: 10.1038/nm917. [DOI] [PubMed] [Google Scholar]
- Harari A, Dutoit V, Cellerai C, Bart PA, Du Pasquier RA, Pantaleo G. Functional signatures of protective antiviral T-cell immunity in human virus infections. Immunol Rev. 2006;211:236–254. doi: 10.1111/j.0105-2896.2006.00395.x. [DOI] [PubMed] [Google Scholar]
- Harari A, Cellerai C, Enders FB, Köstler J, Codarri L, Tapia G, Boyman O, Castro E, Gaudieri S, James I, et al. Skewed association of polyfunctional antigen-specific CD8 T cell populations with HLA-B genotype. Proc Natl Acad Sci USA. 2007;104:16233–16238. doi: 10.1073/pnas.0707570104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz T, Yanover C. Identifying HLA supertypes by learning distance functions. Bioinformatics. 2007;23:e148–e155. doi: 10.1093/Bioinformatics/btl324. [DOI] [PubMed] [Google Scholar]
- Ingram RJ, Chu KK, Metan G, Maillere B, Doganay M, Ozkul Y, Dyson H, Williamson ED, Baillie L, Kim LU, et al. An epitope of Bacillus anthracis protective antigen that is cryptic in rabbits may be immunodominant in humans. Infect Immun. 2010a;78:2353. doi: 10.1128/IAI.00072-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram RJ, Metan G, Maillere B, Doganay M, Ozkul Y, Kim LU, Baillie L, Dyson H, Williamson ED, Chu KK, et al. Natural exposure to cutaneous anthrax gives long-lasting T cell immunity encompassing infection-specific epitopes. J Immunol. 2010b;184:3814–3821. doi: 10.4049/jimmunol.0901581. [DOI] [PubMed] [Google Scholar]
- Jenne L, Hauser C, Arrighi JF, Saurat JH, Hugin AW. Poxvirus as a vector to transduce human dendritic cells for immunotherapy: abortive infection but reduced APC function. Gene Ther. 2000;7:1575–1583. doi: 10.1038/sj.gt.3301287. [DOI] [PubMed] [Google Scholar]
- Jenner E. The Origin of the Vaccines Inoculation. London: Shury; 1801. [Google Scholar]
- Jenner E. An enquiry into the causes and effects of the variolae vaccinae, a disease discovered in some of the western counties of England, particularly Gloucestershire, and known by the name of cow pox. London, 1798. In: Camac CNB, editor. Classics of Medicine and Surgery. New York: Dover; 1959. pp. 213–240. [Google Scholar]
- Jing L, Chong TM, McClurkan CL, Huang J, Story BT, Koelle DM. Diversity in the acute CD8 T cell response to vaccinia virus in humans. J Immunol. 2005;175:7550–7559. doi: 10.4049/jimmunol.175.11.7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing L, Chong TM, Byrd B, McClurkan CL, Huang J, Story BT, Dunkley KM, Aldaz-Carroll L, Eisenberg RJ, Cohen GH, et al. Dominance and diversity in the primary human CD4 T cell response to replication-competent vaccinia virus. J Immunol. 2007;178:6374–6386. doi: 10.4049/jimmunol.178.10.6374. [DOI] [PubMed] [Google Scholar]
- Jing L, Davies DH, Chong TM, Chun S, McClurkan CL, Huang J, Story BT, Molina DM, Hirst S, Felgner PL, Koelle DM. An extremely diverse CD4 response to vaccinia virus in humans is revealed by proteome-wide T-cell profiling. J Virol. 2008;82:7120–7134. doi: 10.1128/JVI.00453-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John M, Heckerman D, James I, Park LP, Carlson JM, Chopra A, Gaudieri S, Nolan D, Haas DW, Riddler SA, et al. Adaptive interactions between HLA and HIV-1: highly divergent selection imposed by HLA class I molecules with common supertype motifs. J Immunol. 2010;184:4368–4377. doi: 10.4049/jimmunol.0903745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kangueane P, Sakharkar MK, Rajaseger G, Bolisetty S, Sivasekari B, Zhao B, Ravichandran M, Shapshak P, Subbiah S. A frame-work to sub-type HLA supertypes. Front Biosci. 2005;10:879–886. doi: 10.2741/1582. [DOI] [PubMed] [Google Scholar]
- Karpenko O, Huang L, Dai Y. A probabilistic meta-predictor for the MHC class II binding peptides. Immunogenetics. 2008;60:25–36. doi: 10.1007/s00251-007-0266-y. [DOI] [PubMed] [Google Scholar]
- Kawashima Y, Pfafferott K, Frater J, Matthews P, Payne R, Addo M, Gatanaga H, Fujiwara M, Hachiya A, Koizumi H, et al. Adaptation of HIV-1 to human leukocyte antigen class I. Nature. 2009;458:641–645. doi: 10.1038/nature07746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenter GG, Welters MJ, Valentijn AR, Lowik MJ, Berends-van der Meer DM, Vloon AP, Essahsah F, Fathers LM, Offringa R, Drijfhout JW, et al. Vaccination against HPV-16 oncoproteins for vulvar intraepithelial neoplasia. N Engl J Med. 2009;361:1838–1847. doi: 10.1056/NEJMoa0810097. [DOI] [PubMed] [Google Scholar]
- Kiepiela P, Ngumbela K, Thobakgale C, Ramduth D, Honeyborne I, Moodley E, Reddy S, de Pierres C, Mncube Z, Mkhwanazi N, et al. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med. 2007;13:46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- Klade CS, Wedemeyer H, Berg T, Hinrichsen H, Cholewinska G, Zeuzem S, Blum H, Buschle M, Jelovcan S, Buerger V, et al. Therapeutic vaccination of chronic hepatitis C nonresponder patients with the peptide vaccine IC41. Gastroenterology. 2008;134:1385–1395. doi: 10.1053/j.gastro.2008.02.058. [DOI] [PubMed] [Google Scholar]
- Koelle DM, Chen HB, Gavin MA, Wald A, Kwok WW, Corey L. CD8 CTL from genital herpes simplex lesions: recognition of viral tegument and immediate early proteins and lysis of infected cutaneous cells. J Immunol. 2001;166:4049–4058. doi: 10.4049/jimmunol.166.6.4049. [DOI] [PubMed] [Google Scholar]
- Koelle DM, Liu Z, McClurkan CL, Cevallos RC, Vieira J, Hosken NA, Meseda CA, Snow DC, Wald A, Corey L. Immunodominance among herpes simplex virus-specific CD8 T cells expressing a tissue-specific homing receptor. Proc Natl Acad Sci USA. 2003;100:12899–12904. doi: 10.1073/pnas.2131705100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koita OA, Dabitao D, Mahamadou I, Tall M, Dao S, Tounkara A, Guiteye H, Noumsi C, Thiero O, Kone M, et al. Confirmation of immunogenic consensus sequence HIV-1 T-cell epitopes in Bamako, Mali and Providence, Rhode Island. Hum Vaccin. 2006;2:119–128. doi: 10.4161/hv.2869. [DOI] [PubMed] [Google Scholar]
- Kosmrlj A, Read EL, Qi Y, Allen TM, Altfeld M, Deeks SG, Pereyra F, Carrington M, Walker BD, Chakraborty AK. Effects of thymic selection of the T-cell repertoire on HLA class I-associated control of HIV infection. Nature. 2010;465:350–354. doi: 10.1038/nature08997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotturi MF, Peters B, Buendia-Laysa F, Jr, Sidney J, Oseroff C, Botten J, Grey H, Buchmeier MJ, Sette A. The CD8+ T-cell response to lymphocytic choriomeningitis virus involves the L antigen: uncovering new tricks for an old virus. J Virol. 2007;81:4928–4940. doi: 10.1128/JVI.02632-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotturi MF, Scott I, Wolfe T, Peters B, Sidney J, Cheroutre H, von Herrath MG, Buchmeier MJ, Grey H, Sette A. Naive precursor frequencies and MHC binding rather than the degree of epitope diversity shape CD8+ T cell immunodominance. J Immunol. 2008;181:2124–2133. doi: 10.4049/jimmunol.181.3.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landais E, Romagnoli PA, Corper AL, Shires J, Altman JD, Wilson IA, Garcia KC, Teyton L. New design of MHC class II tetramers to accommodate fundamental principles of antigen presentation. J Immunol. 2009;183:7949–7957. doi: 10.4049/jimmunol.0902493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauemøller SL, Kesmir C, Corbet SL, Fomsgaard A, Holm A, Claesson MH, Brunak S, Buus S. Identifying cytotoxic T cell epitopes from genomic and proteomic information: “The human MHC project”. Rev Immunogenet. 2000;2:477–491. [PubMed] [Google Scholar]
- Lauer P, Rinaudo CD, Soriani M, Margarit I, Maione D, Rosini R, Taddei AR, Mora M, Rappuoli R, Grandi G, Telford JL. Genome analysis reveals pili in Group B Streptococcus. Science. 2005;309:105. doi: 10.1126/science.1111563. [DOI] [PubMed] [Google Scholar]
- Lehmann PV, Sercarz EE, Forsthuber T, Dayan CM, Gammon G. Determinant spreading and the dynamics of the autoimmune T-cell repertoire. Immunol Today. 1993;14:203–208. doi: 10.1016/0167-5699(93)90163-F. [DOI] [PubMed] [Google Scholar]
- Leslie AJ, Pfafferott KJ, Chetty P, Draenert R, Addo MM, Feeney M, Tang Y, Holmes EC, Allen T, Prado JG, et al. HIV evolution: CTL escape mutation and reversion after transmission. Nat Med. 2004;10:282–289. doi: 10.1038/nm992. [DOI] [PubMed] [Google Scholar]
- Levine MM, Lagos R, Esparza J. Vaccines and Vaccination in Historical Perspective. In: Levine MM, Dougan G, Good MF, Liu MA, Nabel GJ, Nataro JP, Rappuoli R, editors. New Generation Vaccines. Fourth Edition. New York: Informa Healthcare USA, Inc.; 2009. pp. 1–11. [Google Scholar]
- Lewinsohn DA, Winata E, Swarbrick GM, Tanner KE, Cook MS, Null MD, Cansler ME, Sette A, Sidney J, Lewinsohn DM. Immunodominant tuberculosis CD8 antigens preferentially restricted by HLA-B. PLoS Pathog. 2007;3:1240–1249. doi: 10.1371/journal.ppat.0030127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HH, Ray S, Tongchusak S, Reinherz EL, Brusic V. Evaluation of MHC class I peptide binding prediction servers: applications for vaccine research. BMC Immunol. 2008a;9:8. doi: 10.1186/1471-2172-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HH, Zhang GL, Tongchusak S, Reinherz EL, Brusic V. Evaluation of MHC-II peptide binding prediction servers: applications for vaccine research. BMC Bioinformatics. 2008b;9(Suppl 12):S22. doi: 10.1186/1471-2105-9-S12-S22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Xu Z, Fuhlbrigge RC, Peña-Cruz V, Lieberman J, Kupper TS. Vaccinia virus induces strong immunoregulatory cytokine production in healthy human epidermal keratinocytes: a novel strategy for immune evasion. J Virol. 2005;79:7363–7370. doi: 10.1128/JVI.79.12.7363-7370.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund O, Nielsen M, Kesmir C, Petersen AG, Lundegaard C, Worning P, Sylvester-Hvid C, Lamberth K, Røder G, Justesen S, et al. Definition of supertypes for HLA molecules using clustering of specificity matrices. Immunogenetics. 2004;55:797–810. doi: 10.1007/s00251-004-0647-4. [DOI] [PubMed] [Google Scholar]
- Madico G, Welsch JA, Lewis LA, McNaughton A, Perlman DH, Costello CE, Ngampasutadol J, Vogel U, Granoff DM, Ram S. The meningococcal vaccine candidate GNA1870 binds the complement regulatory protein factor H and enhances serum resistance. J Immunol. 2006;177:501–510. doi: 10.4049/jimmunol.177.1.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maecker HT, Dunn HS, Suni MA, Khatamzas E, Pitcher CJ, Bunde T, Persaud N, Trigona W, Fu TM, Sinclair E, et al. Use of overlapping peptide mixtures as antigens for cytokine flow cytometry. J Immunol Methods. 2001;255:27–40. doi: 10.1016/s0022-1759(01)00416-1. [DOI] [PubMed] [Google Scholar]
- Maione D, Margarit I, Rinaudo CD, Masignani V, Mora M, Scarselli M, Tettelin H, Brettoni C, Iacobini ET, Rosini R, et al. Identification of a universal Group B streptococcus vaccine by multiple genome screen. Science. 2005;309:148–150. doi: 10.1126/science.1109869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallios RR. A consensus strategy for combining HLA-DR binding algorithms. Hum Immunol. 2003;64:852–856. doi: 10.1016/s0198-8859(03)00142-3. [DOI] [PubMed] [Google Scholar]
- Manuel ER, Yeh WW, Seaman MS, Furr K, Lifton MA, Hulot SL, Autissier P, Letvin NL. Dominant CD8+ T-lymphocyte responses suppress expansion of vaccine-elicited subdominant T lymphocytes in rhesus monkeys challenged with pathogenic simian-human immunodeficiency virus. J Virol. 2009;83:10028–10035. doi: 10.1128/JVI.01015-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins MA, Wilson NA, Reed JS, Ahn CD, Klimentidis YC, Allison DB, Watkins DI. T-cell correlates of vaccine efficacy after a heterologous simian immunodeficiency virus challenge. J Virol. 2010;84:4352–4365. doi: 10.1128/JVI.02365-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHeyzer-Williams MG, Altman JD, Davis MM. Enumeration and characterization of memory cells in the TH compartment. Immunol Rev. 1996;150:5–21. doi: 10.1111/j.1600-065x.1996.tb00693.x. [DOI] [PubMed] [Google Scholar]
- McMurry J, Sbai H, Gennaro ML, Carter EJ, Martin W, De Groot AS. Analyzing Mycobacterium tuberculosis proteomes for candidate vaccine epitopes. Tuberculosis (Edinb) 2005;85:95–105. doi: 10.1016/j.tube.2004.09.005. [DOI] [PubMed] [Google Scholar]
- McMurry JA, Gregory SH, Moise L, Rivera D, Buus S, De Groot AS. Diversity of Francisella tularensis Schu4 antigens recognized by T lymphocytes after natural infections in humans: identification of candidate epitopes for inclusion in a rationally designed tularemia vaccine. Vaccine. 2007;25:3179–3191. doi: 10.1016/j.vaccine.2007.01.039. [DOI] [PubMed] [Google Scholar]
- Moise L, McMurry JA, Buus S, Frey S, Martin WD, De Groot AS. In silico-accelerated identification of conserved and immunogenic variola/vaccinia T-cell epitopes. Vaccine. 2009;27:6471–6479. doi: 10.1016/j.vaccine.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon JJ, Chu HH, Pepper M, McSorley SJ, Jameson SC, Kedl RM, Jenkins MK. Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity. 2007;27:203–213. doi: 10.1016/j.immuni.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutaftsi M, Peters B, Pasquetto V, Tscharke DC, Sidney J, Bui HH, Grey H, Sette A. A consensus epitope prediction approach identifies the breadth of murine T(CD8+)-cell responses to vaccinia virus. Nat Biotechnol. 2006;24:817–819. doi: 10.1038/nbt1215. [DOI] [PubMed] [Google Scholar]
- Moutaftsi M, Bui HH, Peters B, Sidney J, Salek-Ardakani S, Oseroff C, Pasquetto V, Crotty S, Croft M, Lefkowitz EJ, et al. Vaccinia virus-specific CD4+ T cell responses target a set of antigens largely distinct from those targeted by CD8+ T cell responses. J Immunol. 2007;178:6814–6820. doi: 10.4049/jimmunol.178.11.6814. [DOI] [PubMed] [Google Scholar]
- Moutaftsi M, Salek-Ardakani S, Croft M, Peters B, Sidney J, Grey H, Sette A. Correlates of protection efficacy induced by vaccinia virus-specific CD8+ T-cell epitopes in the murine intranasal challenge model. Eur J Immunol. 2009;39:717–722. doi: 10.1002/eji.200838815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutaftsi M, Tscharke DC, Vaughan K, Koelle DM, Stern L, Calvo-Calle M, Ennis F, Terajima M, Sutter G, Crotty S, et al. Uncovering the interplay between CD8, CD4 and antibody responses to complex pathogens. Future Microbiol. 2010;5:221–239. doi: 10.2217/fmb.09.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynihan JS, Howard CR. Recent advances in the development of peptide vaccines for hepatitis B. Intervirology. 2001;44:65–77. doi: 10.1159/000050034. [DOI] [PubMed] [Google Scholar]
- Nielsen M, Lundegaard C, Blicher T, Lamberth K, Harndahl M, Justesen S, Røder G, Peters B, Sette A, Lund O, Buus S. NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence. PLoS ONE. 2007;2:e796. doi: 10.1371/journal.pone.0000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M, Lundegaard C, Blicher T, Peters B, Sette A, Justesen S, Buus S, Lund O. Quantitative predictions of peptide binding to any HLA-DR molecule of known sequence: NetMHCIIpan. PLoS Comput Biol. 2008;4:e1000107. doi: 10.1371/journal.pcbi.1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M, Lund O, Buus S, Lundegaard C. MHC Class II epitope predictive algorithms. Immunology. 2010;130:319–328. doi: 10.1111/j.1365-2567.2010.03268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offit PA. Vaccinated: One Man’s Quest to Defeat the World’s Deadliest Diseases. New York: HarperCollins; 2007. [Google Scholar]
- Oseroff C, Kos F, Bui HH, Peters B, Pasquetto V, Glenn J, Palmore T, Sidney J, Tscharke DC, Bennink JR, et al. HLA class I-restricted responses to vaccinia recognize a broad array of proteins mainly involved in virulence and viral gene regulation. Proc Natl Acad Sci USA. 2005;102:13980–13985. doi: 10.1073/pnas.0506768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou D, Mitchell LA, Tingle AJ. A new categorization of HLA DR alleles on a functional basis. Hum Immunol. 1998;59:665–676. doi: 10.1016/s0198-8859(98)00067-6. [DOI] [PubMed] [Google Scholar]
- Panchanathan V, Chaudhri G, Karupiah G. Correlates of protective immunity in poxvirus infection: where does antibody stand? Immunol Cell Biol. 2008;86:80–86. doi: 10.1038/sj.icb.7100118. [DOI] [PubMed] [Google Scholar]
- Pasteur L. De l’attenuation du virus du Choléra des poules. CR Acad Sci Paris. 1880;91:673–680. [Google Scholar]
- Peters B, Bui HH, Frankild S, Nielson M, Lundegaard C, Kostem E, Basch D, Lamberth K, Harndahl M, Fleri W, et al. A community resource benchmarking predictions of peptide binding to MHC-I molecules. PLoS Comput Biol. 2006;2:e65. doi: 10.1371/journal.pcbi.0020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizza M, Scarlato V, Masignani V, Giuliani MM, Aricò B, Comanducci M, Jennings GT, Baldi L, Bartolini E, Capecchi B, et al. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science. 2000;287:1816–1820. doi: 10.1126/science.287.5459.1816. [DOI] [PubMed] [Google Scholar]
- Provenzano M, Selleri S, Jin P, Wang E, Werden R, Slezak S, Adams SD, Panelli MC, Leitman SF, Stroncek DF, Marincola FM. Comprehensive epitope mapping of the Epstein-Barr virus latent membrane protein-2 in normal, non tumor-bearing individuals. Cancer Immunol Immunother. 2007;56:1047–1063. doi: 10.1007/s00262-006-0246-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putz MM, Alberini I, Midgley CM, Manini I, Montomoli E, Smith GL. Prevalence of antibodies to Vaccinia virus after smallpox vaccination in Italy. J Gen Virol. 2005;86:2955–2960. doi: 10.1099/vir.0.81265-0. [DOI] [PubMed] [Google Scholar]
- Ramon G. Sur la toxine et sur l’anatoxine diphtériques. Pouvoir floculant et propriétés immunisantes. Ann Inst Pasteur (Paris) 1924;38:1–106. [Google Scholar]
- Rappuoli R. Reverse vaccinology. Curr Opin Microbiol. 2000;3:445–450. doi: 10.1016/s1369-5274(00)00119-3. [DOI] [PubMed] [Google Scholar]
- Rappuoli R. From Pasteur to genomics: progress and challenges in infectious diseases. Nat Med. 2004;10:1177–1185. doi: 10.1038/nm1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reche PA, Reinherz EL. Definition of MHC supertypes through clustering of MHC peptide binding repertoires. In: Nicosia G, Cutello V, Bentley PJ, Timmis T, editors. Proceedings of 3rd International Conference on Artificial Immune Systems. ICARIS 2004, LNCS 3239. Heidelberg: Springer-Verlag; 2004. pp. 189–196. [Google Scholar]
- Robb ML. Failure of the Merck HIV vaccine: an uncertain step forward. Lancet. 2008;372:1857–1858. doi: 10.1016/S0140-6736(08)61593-7. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Ortega MJ, Norais N, Bensi G, Liberatori S, Capo S, Mora M, Scarselli M, Doro F, Ferrari G, Garaguso I, et al. Characterization and identification of vaccine candidate proteins through analysis of the group A Streptococcus surface proteome. Nat Biotechnol. 2006;24:191–197. doi: 10.1038/nbt1179. [DOI] [PubMed] [Google Scholar]
- Santra S, Liao HX, Zhang R, Muldoon M, Watson S, Fischer W, Theiler J, Szinger J, Balachandran H, Buzby A, et al. Mosaic vaccines elicit CD8+ T lymphocyte responses that confer enhanced immune coverage of diverse HIV strains in monkeys. Nat Med. 2010;16:324–328. doi: 10.1038/nm.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider MC, Prosser BE, Caesar JJ, Kugelberg E, Li S, Zhang Q, Quoraishi S, Lovett JE, Deane JE, Sim RB, et al. Neisseria meningitidis recruits factor H using protein mimicry of host carbohydrates. Nature. 2009;458:890–893. doi: 10.1038/nature07769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sester U, Presser D, Dirks J, Gärtner BC, Köhler H, Sester M. PD-1 expression and IL-2 loss of cytomegalovirus- specific T cells correlates with viremia and reversible functional anergy. Am J Transplant. 2008;8:1486–1497. doi: 10.1111/j.1600-6143.2008.02279.x. [DOI] [PubMed] [Google Scholar]
- Sette A, Sidney J. HLA supertypes and supermotifs: a functional perspective on HLA polymorphism. Curr Opin Immunol. 1998;10:478–482. doi: 10.1016/s0952-7915(98)80124-6. [DOI] [PubMed] [Google Scholar]
- Sette A, Sidney J. Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism. Immunogenetics. 1999;50:201–212. doi: 10.1007/s002510050594. [DOI] [PubMed] [Google Scholar]
- Sette A, Moutaftsi M, Moyron-Quiroz J, McCausland MM, Davies DH, Johnston RJ, Peters B, Rafii-El-Idrissi Benhnia M, Hoffmann J, Su HP, et al. Selective CD4+ T cell help for antibody responses to a large viral pathogen: deterministic linkage of specificities. Immunity. 2008;28:847–858. doi: 10.1016/j.immuni.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidney J, Peters B, Frahm N, Brander C, Sette A. HLA class I supertypes: a revised and updated classification. BMC Immunol. 2008;9:1. doi: 10.1186/1471-2172-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder EE, Kampanya N, Lu J, Nordberg EK, Karur HR, Shukla M, Soneja J, Tian Y, Xue T, Yoo H, et al. PATRIC: the VBI Patho-Systems Resource Integration Center. Nucleic Acids Res. 2007;35(Database issue):D401–D406. doi: 10.1093/nar/gkl858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwood S, Sidney J, Kondo A, del Guercio MF, Appella E, Hoffman S, Kubo RT, Chesnut RW, Grey HM, Sette A. Several common HLA-DR types share largely overlapping peptide binding repertoires. J Immunol. 1998;160:3363–3373. [PubMed] [Google Scholar]
- Squires B, Macken C, Garcia-Sastre A, Godbole S, Noronha J, Hunt V, Chang R, Larsen CN, Klem E, Biersack K, Scheuermann RH. BioHealthBase: informatics support in the elucidation of influenza virus host pathogen interactions and virulence. Nucleic Acids Res. 2008;36(Database issue):D497–D503. doi: 10.1093/nar/gkm905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H, Saunders NJ, Heidelberg J, Jeffries AC, Nelson KE, Eisen JA, Ketchum KA, Hood DW, Peden JF, Dodson RJ, et al. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science. 2000;287:1809–1815. doi: 10.1126/science.287.5459.1809. [DOI] [PubMed] [Google Scholar]
- Thorpe C, Edwards L, Snelgrove R, Finco O, Rae A, Grandi G, Guilio R, Hussell T. Discovery of a vaccine antigen that protects mice from Chlamydia pneumoniae infection. Vaccine. 2007;25:2252–2260. doi: 10.1016/j.vaccine.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Tobery TW, Wang S, Wang XM, Neeper MP, Jansen KU, McClements WL, Caulfield MJ. A simple and efficient method for the monitoring of antigen-specific T cell responses using peptide pool arrays in a modified ELISpot assay. J Immunol Methods. 2001;254:59–66. doi: 10.1016/s0022-1759(01)00397-0. [DOI] [PubMed] [Google Scholar]
- Toebes M, Coccoris M, Bins A, Rodenko B, Gomez R, Nieuwkoop NJ, van de Kasteele W, Rimmelzwaan GF, Haanen JB, Ovaa H, Schumacher TN. Design and use of conditional MHC class I ligands. Nat Med. 2006;12:246–251. doi: 10.1038/nm1360. [DOI] [PubMed] [Google Scholar]
- Tong JC, Tan TW, Ranganathan S. In silico grouping of peptide/HLA class I complexes using structural interaction characteristics. Bioinformatics. 2007;23:177–183. doi: 10.1093/bioinformatics/btl563. [DOI] [PubMed] [Google Scholar]
- Leisner C, Loeth N, Lamberth K, Justesen S, Sylvester-Hvid C, Schmidt EG, Claesson M, Buus S, Stryhn A. One-pot, mix-and-read peptide-MHC tetramers. PLoS ONE. 2008;3:e1678. doi: 10.1371/journal.pone.0001678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan K, Blythe M, Greenbaum J, Zhang Q, Peters B, Doolan DL, Sette A. Meta-analysis of immune epitope data for all Plasmodia: overview and applications for malarial immunobiology and vaccine-related issues. Parasite Immunol. 2009;31:78–97. doi: 10.1111/j.1365-3024.2008.01077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh SR, Gillis J, Peters B, Mothé BR, Sidney J, Sette A, Johnson RP. Diverse recognition of conserved orthopoxvirus CD8+ T cell epitopes in vaccinated rhesus macaques. Vaccine. 2009;27:4990–5000. doi: 10.1016/j.vaccine.2009.05.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Sidney J, Dow C, Mothé B, Sette A, Peters B. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput Biol. 2008;4:e1000048. doi: 10.1371/journal.pcbi.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson NA, Keele BF, Reed JS, Piaskowski SM, MacNair CE, Bett AJ, Liang X, Wang F, Thoryk E, Heidecker GJ, et al. Vaccine-induced cellular responses control simian immunodeficiency virus replication after heterologous challenge. J Virol. 2009;83:6508–6521. doi: 10.1128/JVI.00272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu RH, Remakus S, Ma X, Roscoe F, Sigal LJ. Direct presentation is sufficient for an efficient anti-viral CD8+ T cell response. PLoS Pathog. 2010;6:e1000768. doi: 10.1371/journal.ppat.1000768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauch LE, Zellweger RM, Kotturi MF, Qutubuddin A, Sidney J, Peters B, Prestwood TR, Sette A, Shresta S. A protective role for dengue virus-specific CD8+ T cells. J Immunol. 2009;182:4865–4873. doi: 10.4049/jimmunol.0801974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yewdell JW, Nicchitta CV. The DRiP hypothesis decennial: support, controversy, refinement and extension. Trends Immunol. 2006;27:368–373. doi: 10.1016/j.it.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Wang P, Kim Y, Haste-Andersen P, Beaver J, Bourne PE, Bui HH, Buus S, Frankild S, Greenbaum J, et al. Immune epitope database analysis resource (IEDB-AR) Nucleic Acids Res. 2008;36:W513–W518. doi: 10.1093/nar/gkn254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerli SC, Harari A, Cellerai C, Vallelian F, Bart PA, Pantaleo G. HIV-1-specific IFN-gamma/IL-2-secreting CD8 T cells support CD4-independent proliferation of HIV-1-specific CD8 T cells. Proc Natl Acad Sci USA. 2005;102:7239–7244. doi: 10.1073/pnas.0502393102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuñiga R, Lucchetti A, Galvan P, Sanchez S, Sanchez C, Hernandez A, Sanchez H, Frahm N, Linde CH, Hewitt HS, et al. Relative dominance of Gag p24-specific cytotoxic T lymphocytes is associated with human immunodeficiency virus control. J Virol. 2006;80:3122–3125. doi: 10.1128/JVI.80.6.3122-3125.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]


