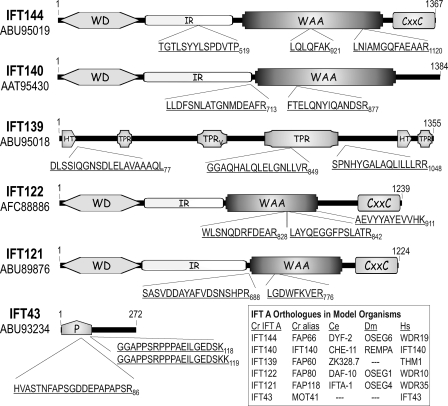
FIGURE 1.
Chlamydomonas IFT A protein analysis. Sucrose density gradient-purified IFT A proteins were separated by electrophoresis with isolated tryptic peptides analyzed by Edman degradation as described under “Experimental Procedures.” IFT43 was excised from an SDS-PAGE gel of immunopurified IFT A (see supplemental Fig. S1) prior to trypsinization and MALDI-TOF mass spectroscopy. Three peptide masses resulting from the IFT43 analysis, 2122.9794, 1874.9612, and 2003.0562 Da, matched predicted masses of three peptide sequences, HVASTNFAPSGDDEPAPAPSR86, GGAPPSRPPPAEILGEDSK118, and GGAPPSRPPPAEILGEDSKK119, respectively, that came from a single ORF in the Chlamydomonas genome. The third IFT43 peptide resulted from an incomplete cleavage at Lys119. WD, WD repeat domain; IR, intervening region; WAA, degenerate TPR-like repeats; CXXC, Cys-Xaa-Xaa-Cys repeat domain; TPR, tetratricopeptide repeats; TPRV, TPR domain present in vertebrate THM1/IFT139; HT, half of a tetratricopeptide repeat; P, proline-rich domain.