Background: It remains unknown whether the spindle assembly checkpoint (SAC) also requires the nonkinetochore protein.
Results: RED localizes to the spindle poles, determines kinetochore localization of MAD1, and exerts its effect on the SAC.
Conclusion: The SAC also requires the spindle pole-associated protein.
Significance: The kinetochore protein coordinates with the nonkinetochore protein for activation of the SAC.
Keywords: Cell Biology, Cell Cycle, Checkpoint Control, Kinetochore, Mitosis
Abstract
The spindle assembly checkpoint (SAC) is essential for ensuring the proper attachment of kinetochores to the spindle and, thus, the precise separation of paired sister chromatids during mitosis. The SAC proteins are recruited to the unattached kinetochores for activation of the SAC in prometaphase. However, it has been less studied whether activation of the SAC also requires the proteins that do not localize to the kinetochores. Here, we show that the nuclear protein RED, also called IK, a down-regulator of human leukocyte antigen (HLA) II, interacts with the human SAC protein MAD1. Two RED-interacting regions identified in MAD1 are from amino acid residues 301–340 and 439–480, designated as MAD1(301–340) and MAD1(439–480), respectively. Our observations reveal that RED is a spindle pole-associated protein that colocalizes with MAD1 at the spindle poles in metaphase and anaphase. Depletion of RED can cause a shorter mitotic timing, a failure in the kinetochore localization of MAD1 in prometaphase, and a defect in the SAC. Furthermore, the RED-interacting peptides MAD1(301–340) and MAD1(439–480), fused to enhanced green fluorescence protein, can colocalize with RED at the spindle poles in prometaphase, and their expression can abrogate the SAC. Taken together, we conclude that RED is required for kinetochore localization of MAD1, mitotic progression, and activation of the SAC.
Introduction
In eukaryotic cells, the spindle assembly checkpoint (SAC)2 can monitor the integrity of the mitotic spindle and the attachment of paired sister chromatids to the spindle, thereby ensuring precise separation of paired sister chromatids into two daughter cells during mitosis (1). The core components of the human SAC are thought to include Aurora B, MPS1, MAD1, MAD2, BUB1, BUBR1, BUB3, and CDC20 (1). The recruitment of these checkpoint proteins to the unattached kinetochore eventually induce the formation of an active form of MAD2, which can inhibit CDC20, an activator of the anaphase-promoting complex/cyclosome (APC/C) that functions as a ubiquitin ligase for proteolysis of securin and cyclin B1 (1). When paired sister chromatids are properly attached to the spindle microtubules and aligned along the metaphase plate, the MAD1-MAD2 complexes are released from the spindle-attached kinetochores and relocalized to the spindle poles for silencing of the SAC (2–4). Upon inactivation of the SAC, CMT2/p31Comet activates the APCCDC20, leading to proteolysis of securin and cyclin B1 (5–8). Securin proteolysis releases separase to cleave cohesin, which binds together paired sister chromatids (9, 10). Digestion of cohesin results in the separation of paired sister chromatids to promote entry into anaphase (11).
Human MAD1 was originally identified by yeast two-hybrid screening as a protein interacting with the human T cell leukemia virus type 1 oncoprotein Tax (12). Binding of Tax to MAD1 can cause a defect in the SAC (12). MAD1 interacts with and is required to recruit MAD2 to kinetochores during prometaphase or in response to spindle damage (12, 13). To identify the proteins that interact with human MAD1, we performed yeast two-hybrid screening and obtained a gene encoding RED (also known as IK), a nuclear protein of 557 amino acids (14–17). The term RED is derived from its sequence, which contains an extensive stretch of alternating arginine (R) and glutamic acid (E) or aspartic acid (D) residues from amino acid residues 334 to 375 (14). Some studies have shown that RED has an inhibitory role in regulating Human Leukocyte Antigen (HLA) class II gene expression and that RED associates with the spliceosome complex (15, 16). In this study, we found that the protein levels of RED increased from early S to M phases along with cyclin B1 and securin. This prompted us to further investigate the role of RED in the SAC and in mitosis. Here we provide evidence to demonstrate that RED has a role in the activation of the SAC and in mitosis progression.
EXPERIMENTAL PROCEDURES
Plasmid Constructs
Full-length MAD1 cDNA was cloned into the Gal4BD (DNA-binding domain of Gal4) vector pBDGAL4 CAM (Stratagene) for yeast two-hybrid screening. Full-length MAD1 and RED cDNAs, or cDNA fragments, were cloned into the Gal4BD vector pAS2-1 (Clontech) and the Gal4AD (activation domain of Gal4) vector pACT2 (Clontech Co.) or pADGAL4 (Stratagene) for yeast two-hybrid analyses. To identify the RED-interacting region in MAD1 by yeast two-hybrid analysis, MAD1 deletion mutants (MAD1(576–718), amino acids (aa) 576–718; MAD1(1–480), aa 1–480; MAD1(1–340), aa 1–340; MAD1(1–160), aa 1–160; MAD1(232–340), aa 232–340; MAD1(301–340), aa 301–340; and MAD1(439–480), aa 439–480) were fused in-frame with Gal4BD in the pAS2-1 vector. Also, MAD1(301–340) was fused in-frame with Gal4AD in the pADGAL4 vector.
The full-length MAD1, MAD1(232–340), MAD1(301–340), and MAD1(439–480) DNA fragments were cloned into the pGEX-KG vector (Amersham Biosciences) for expression of GST-MAD1, GST-MAD1(232–340), GST-MAD1(301–340), and GST-MAD1(439–480) in Escherichia coli strain BL21, respectively. The MAD1(1–160) DNA fragment was cloned into both pET32a (Novagen) and pGEX-KG vectors for overexpression of GST-tagged and His-tagged MAD1(1–160) recombinant proteins in E. coli strain BL21(DE3). GST-tagged and His-tagged MAD1(1–160) recombinant proteins were used for the production and purification of rat anti-MAD1 antibodies, respectively.
The full-length RED was cloned into pB479 (a modified vector of pET11d obtained from Dr. Pellman, Dana-Farber Cancer Institute) for overexpression of His-HA-tagged RED (His-HA-RED) in E. coli strain BL21(DE3). RED(1–115), a DNA fragment encoding the amino acid residues 1 to 115 of RED, was cloned into pGEX-KG and pET30b (Novagen) for overexpression of GST-tagged and His-tagged RED(1–115) recombinant proteins in E. coli, respectively, which were used for the production and purification of rabbit anti-RED antibodies.
For moderate expression of EGFP (enhanced green fluorescence protein of Aequorea victoria) and its fusion genes in cells, the partially disabled encephalomyocarditis virus internal ribosome entry sequence (IRES) (18) from pIRES2-AcGFP (Clontech) and the EGFP DNA sequence from pEGFP-C1 (Clontech) were recombined and subcloned into the pCDNA3 vector (Invitrogen). The resulting construct was referred to as pIRES-EGFP. For comparison, EGFP was also subcloned into pCDNA3 to generate a pEGFP vector for overexpression of EGFP and its fusion genes. Full-length MAD1, MAD1(301–340), and MAD1(439–480) DNA fragments were cloned into the pIRES-EGFP vector to obtain pIRES-EGFP-MAD1, pIRES-EGFP-MAD1(301–340), and pIRES-EGFP-MAD1(439–480), respectively. Full-length MAD1 was also cloned into the pEGFP vector to obtain pEGFP-MAD1.
The full-length wild-type RED was cloned into pEGFP-C2 (Clontech) to generate pEGFP-REDwt. We also cloned REDRES, a rescue allele of RED that is resistant to the RED siRNA#1 duplex, into pEGFP-C2 to generate pEGFP-REDRES. The target sequence of the RED siRNA#1 duplex in wild-type RED is 5′-AGACCACACTGACCACAAA-3′ and was altered to 5′-AGACCACCTTAACTACTAA-3′ in REDRES. The altered nucleotide sequence in REDRES did not change the amino acid sequence of RED.
Yeast Two-hybrid Screening
Yeast two-hybrid screening was performed following the manufacturer's protocol (Clontech). The GAL4BD-MAD1 bait vector and a human liver cDNA library (HL4029AH, Clontech) cloned into the ADGAL4 vector pACT2 were transformed into the yeast YRG-2 strain (Stratagene) for screening. Yeast transformants were directly spread and screened on synthetic dextrose minimal solid medium lacking leucine, tryptophan, and histidine and containing 25 mm 3-aminotriazole to assay for the expression of the HIS3 reporter gene. The screened transformants were further examined by performing an 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) colony filter assay to detect the expression of the lacZ reporter gene, as described in the manual (Clontech).
Antibody Production
Purified His-tagged MAD1(1–160) and His-tagged RED(1–115) recombinant proteins were mixed with Freund's adjuvant (Sigma) to generate polyclonal antibodies against MAD1 and RED, respectively. Rats were immunized by repeated subcutaneous injections with 100 μg of His-tagged MAD1(1–160), and rabbits were similarly immunized with 150 μg of His-tagged RED(1–115) proteins. Rabbit anti-RED antibodies were further purified using Affi-Gel 10 beads conjugated with GST-RED(1–115) recombinant proteins following the standard procedure as described in the manufacturer's manual (Bio-Rad). Likewise, rat anti-MAD1 antibodies were further purified using Affi-Gel 10 beads conjugated with GST-MAD1(1–160) recombinant proteins. The purified antibodies were dialyzed with 1× PBS buffer containing 50% glycerol and 2 mm 2-mercaptoethanol for storage.
Cell Culture, Transfections, and Cell Synchronization
HeLa and H2B-GFP HeLa cells were used in this study. Cells were cultivated following standard procedures. Cell transfection was performed using Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol. For each transfection, 300 ng of plasmid DNA was used. To arrest cells in early S phase, cells were treated with a double thymidine (2 mm) block. To arrest cells in M phase, cells were treated with 50 ng/ml nocodazole for 18 h.
In Vitro Binding Assay and Coimmunoprecipitation
To perform in vitro GST pull-down assays, overexpressed GST, GST-MAD1, GST-MAD1(232–340), GST-MAD1(301–340), GST-MAD1(439–480), and His-HA-tagged RED (His-HA-RED) proteins were purified from E. coli BL21(DE3) according to the manufacturers' protocols (Novagen and Amersham Biosciences). To perform in vitro binding assays, GST (∼1.5 μg) or GST-MAD1 (∼0.24 μg) and His-HA-RED (∼0.12 μg) were incubated in 1 ml of RIPA buffer (50 mm Tris-HCl (pH 7.4), 0.1% Nonidet P-40, 150 mm NaCl, 2 mm DTT) at 4 °C for 1 h. GST or GST-MAD1 was then precipitated using glutathione-Sepharose beads. The precipitated proteins were separated by SDS-PAGE and analyzed by immunoblot analysis.
To perform in vitro binding assays using EGFP-MAD1 fusion proteins, EGFP or EGFP-MAD1 proteins were affinity-purified from HeLa cells stably transfected with pIRES-EGFP or pIRES-EGFP-MAD1 using GFP-Trap®_A beads (Chromotek Co.). The purified EGFP or EGFP-MAD1 proteins were kept bound to the beads and washed with wash buffer (50 mm Tris-HCl (pH 7.4), 1% Nonidet P-40, 1 m NaCl, 2 mm DTT) three times before the beads were resuspended in RIPA buffer containing 50% glycerol for storage. To determine the MAD1-RED interaction, EGFP (∼40 ng) or EGFP-MAD1 (∼160 ng) and His-HA-RED (∼50 ng) were incubated in 1 ml of RIPA buffer (50 mm Tris-HCl (pH 7.4), 0.1% Nonidet P-40, 150 mm NaCl, 2 mm DTT) at 4 °C for 1 h. The bound proteins were spun down, separated by SDS-PAGE, and analyzed by immunoblot analysis.
Immunoprecipitation assays were also performed in RIPA buffer containing a protease inhibitor mixture (Roche) and phosphatase inhibitors (Sigma) using rabbit anti-RED (made in this study) or rabbit anti-MAD1 (Genetex Co.) antibodies in combination with protein G beads (Amersham Biosciences) or using GFP-Trap®_A beads (Chromotek Co.) alone. The proteins precipitated from the cell lysates were analyzed as described above.
RNA Interference Assay
RED siRNA#1 and #2 duplexes were used to deplete RED in HeLa cells. The sense strand sequences of these two duplexes were 5′-r(AGACCACACUGACCACAAA)dTdT-3′ and 5′-r(AGCUGAGAUUGCCAGCAAA)dTdT-3′, respectively. The sense strand sequence of the MAD1 siRNA duplex used in this study was 5′-r(CAGGCAGUGUCAGCAGAAC)dTdT-3′ (19), whereas the sense strand sequence of the non-silencing siRNA duplex was 5′-r(UUCUCCGAACGUGUCACGU)dTdT-3′. All siRNA duplexes were chemically synthesized by and purchased from Qiagen Co. Cells were transfected with 200 pmols of siRNA duplex using oligofectamine as described in the manufacturer's protocol (Invitrogen). To rescue the defect in the SAC caused by the RED siRNA#1-directed depletion of RED, 100 pmols of RED siRNA#1 duplex and 150 ng of pEGFP-REDRES plasmid DNA were cotransfected into cells using Lipofectamine 2000 following the manufacturer's protocol (Invitrogen Co.).
Immunoblot Analysis and Immunofluorescence Microscopy
To perform immunoblot analysis, cells were lysed with RIPA buffer, and cell lysates were resolved using 10% or 12% SDS-PAGE and transferred to PVDF membranes. RED, MAD1, MAD2, Cyclin B1, Securin, β-tubulin, and GFP were detected with their specific antibodies (rabbit anti-RED and rat anti-MAD1 antibodies made in this study, mouse anti-MAD2 antibody from Abnova Co. or Genetex Co., rabbit anti-securin antibody from Zymed Laboratories Inc., mouse anti-securin antibody from Abcam, mouse anti-β-tubulin antibody from Sigma-Aldrich, and mouse anti-GFP antibody from Sigma-Aldrich) and visualized by chemiluminescence (PerkinElmer Life Sciences) after incubation with HRP-conjugated secondary antibodies.
Immunofluorescence staining for MAD1 and RED were performed simultaneously as modified from Campbell et al. (2001) (20). Cells were washed twice with 1 ml of PHEM buffer (60 mm PIPES, 25 mm HEPES (pH 6.9), 10 mm EGTA, and 4 mm MgCl2) and fixed with 1% paraformaldehyde in PHEM buffer. Cells were further permeabilized with 1% CHAPS in PHEM buffer before being treated with filtered MBST blocking buffer (10 mm MOPS (pH 7.4), 150 mm NaCl, 0.05% Tween 20) containing 1% skim milk. Purified rat anti-MAD1 (1 mg/ml) and rabbit anti-RED antibodies (0.5 mg/ml) were diluted 1/150 and 1/50, respectively, in filtered MBST with 1% skim milk. FITC and Cy3-conjugated secondary antibodies (Jackson ImmunoResearch Laboratory) were diluted 1/25 and 1/100, respectively, in filtered MBST with 1% skim milk. To stain EGFP-MAD1 and its derivatives, chicken anti-GFP antibodies (Abcam) were used at a dilution of 1/250 in filtered MBST with 1% skim milk. To stain kinetochores, human anti-centromere antibodies (Sigma-Aldrich) were used at a dilution of 1/100 in filtered MBST with 1% skim milk. DAPI was used to stain chromosomal DNA.
To stain RED and γ-tubulin simultaneously, cells were rinsed twice with PBS, fixed using cold methanol, and blocked using filtered PBS buffer with 1% skim milk. Purified rabbit anti-RED antibodies and mouse monoclonal anti-γ-tubulin sera were diluted 1/50 and 1/500, respectively, in filtered PBS buffer with 1% skim milk. FITC and Cy3-conjugated secondary antibodies were diluted 1/50 and 1/500, respectively, in filtered PBS with 1% skim milk. All of the antibody incubations were carried out for 1 h at room temperature, followed by three washes with PBS buffer.
All stained cells were visualized using either an E800 fluorescence microscope (Nikon Instech) or a confocal laser microscope (Leica). Red and green fluorescence signals were merged using computer software (Leica).
Live Cell Imaging
To perform live cell analysis, H2B-GFP HeLa cells (obtained from Dr. Wahl, The Salk Institute, San Diego, CA) were cultured on poly-l-lysine-coated cover slides and imaged every 5 min using a Zeiss inverted fluorescence microscope (Axio Observer Z1) equipped with the CoolSNAP HQ monochrome camera (Photometrics). All images were analyzed with Axiovision imaging software (release 4.8.2, Carl Zeiss).
Flow Cytometry
Analysis for the DNA content of cells was performed as modified from Rogers (21). Cells were harvested, washed with cold PBS buffer, and fixed in 5 ml of −20 °C ethanol. The fixed cells were spun down, washed three times with PBS buffer containing 1% BSA, and resuspended in 0.8 ml PBS containing 1% BSA. Propidium iodide was added to a final concentration of 20 μg/ml to stain DNA, and RNase A was added to a final concentration of 1 mg/ml to digest RNA. Propidium iodide-stained cells were analyzed on a FACScanTM using CellQuest software (BD Biosciences).
Statistical Analysis
Multiple statistical comparisons (Fisher Least Significant Difference Method, α = 0.05) were performed with analysis of variance tests using Sigma Stat 2.03 software.
RESULTS
RED Is Identified as a MAD1-interacting Protein by Yeast Two-hybrid Screening
We performed yeast two-hybrid screening to identify the proteins that interact with MAD1 in human cells. Full-length MAD1 was fused in-frame to the DNA-BD of the yeast Gal4 protein and used as a bait to screen a human liver cDNA library that was fused to the yeast Gal4 AD (see “Experimental Procedures”). From a total of ∼1 × 106 transformants, seven proteins were identified as being able to interact with MAD1, including RED, CENP-F, CEP250, ZAP113, fibrinogen, LMO7, and p116Rip. For unknown reasons, MAD2 was not identified in our screen using this human liver cDNA library. Nevertheless, CENP-F has been identified previously as a MAD1-interacting protein in the yeast two-hybrid screen that was conducted by Martin-Lluesma et al. (see the supplement in Ref. 13). Of these seven proteins, CENP-F is a kinetochore protein that has been shown to regulate the kinetochore localization of MAD1 and has an essential role in the SAC (22); CEP250, a centrosomal protein, has a role in centrosome cohesion (23). Because RED is a nuclear protein, as is MAD1 (14, 20), we chose to study it further (see yeast two-hybrid analysis for the MAD1-RED interaction in supplemental Fig. S1).
RED Expression Is Cell Cycle-regulated
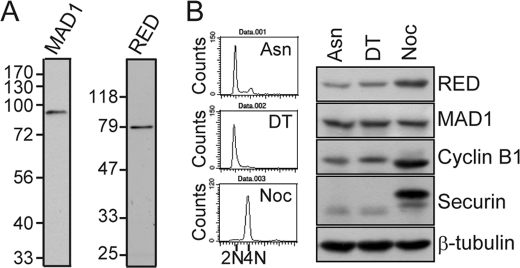
To characterize the RED protein, we purified the bacterially expressed recombinant protein representing the 115 N-terminal amino acid residues of RED, RED(1–115), and used it to raise rabbit antibodies against RED. Likewise, the recombinant protein representing the 160 N-terminal amino acid residues of MAD1, MAD1(1–160), was used to raise rat antibodies against MAD1. The antibodies were affinity-purified (see “Experimental Procedures”), and each one recognized a single protein band in immunoblot analyses (Fig. 1A). Furthermore, peptide blocking tests also showed that preincubation of the antibodies with the RED(1–115) or MAD1(1–160) recombinant peptides could abolish the appearance of the indicated signals representing RED and MAD1 by immunoblot analysis, respectively (data not shown), revealing the specificities of the antibodies. The aberrant electrophoretic migration of RED (65 kDa) observed here was consistent with previous observations (14), and the electrophoretic migration of MAD1 was consistent with its calculated molecular weight (83 kDa) (20).
FIGURE 1.
Antibodies against MAD1 and RED (A). Asynchronous HeLa cell lysates were subjected to SDS-PAGE and processed for Western blotting using an affinity-purified rat anti-MAD1 or rabbit anti-RED antibody as a probe. B, protein levels of RED in early S and M phases. Cells were treated with a double thymidine (DT; thymidine, 2 mm) block to arrest them in early S phase or with nocodazole (Noc, 50 ng/ml) to arrest them in M phase. Nocodazole-arrested mitotic cells were shaken off from the Petri dish and collected for Western blot analysis. Asynchronous cell lysates (Asn) were used for comparison. Fifty micrograms of cell lysates were used for Western blotting with antibodies against RED, MAD1, cyclin B1, securin, and β-tubulin (loading control). Cells were also collected, fixed with cold ethanol, and stained with propidium iodide to determine DNA content by flow cytometric analysis.
To determine whether the protein levels of RED vary during cell cycle progression, we examined the amount of RED in different phases of the cell cycle. Cells were treated with a double thymidine block to arrest them in the early S phase and with nocodazole, a microtubule destabilizer, to arrest them in the M phase. The DNA content of the early S-phase and M-phase cells was 2N and 4N, respectively, as confirmed by flow cytometric analysis (Fig. 1B). β-tubulin was used as an internal control to compare the amount of RED, MAD1, cyclin B1, and securin in the early S- and M-phase cells (Fig. 1B). Unlike MAD1 (20), the protein levels of RED increased from the early S to M phase, along with cyclin B1 and securin, indicating that RED expression is cell cycle-regulated and implying that RED has a role in mitosis.
MAD1 Binds to RED in Vitro and in Vivo
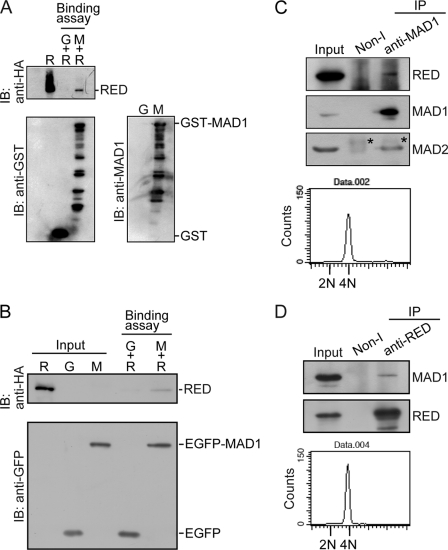
To confirm the interaction between MAD1 and RED in vitro, a protein precipitation assay with glutathione beads was performed using a GST fusion protein. We found that the GST-MAD1 fusion proteins were able to interact with the bacterially expressed RED proteins, whereas GST could not (Fig. 2A). Considering that GST-MAD1 proteins were severely degraded when expressed in bacterial cells (Fig. 2A), we further affinity-purified the EGFP-MAD1 fusion proteins from the EGFP-MAD1-expressing HeLa cells using beads conjugated with anti-GFP antibodies (see “Experimental Procedures”) for the in vitro binding assay. Consistently, the affinity-purified EGFP-MAD1 fusion proteins were also able to interact with the bacterially expressed RED proteins (Fig. 2B). Therefore, these results indicate that MAD1 may bind directly to RED.
FIGURE 2.
MAD1 binds to RED. A and B, MAD1 binds to RED in vitro. A, purified GST-MAD1 interacts with His-HA-RED. Purified GST (G) or GST-MAD1 (M) recombinant fusion proteins were incubated with purified His-HA-RED (R) recombinant proteins in RIPA buffer and precipitated with glutathione-Sepharose beads at 4 °C. The precipitated proteins were analyzed by immunoblot (IB). Purified His-HA-RED (R) recombinant protein was loaded as an input control. B, affinity-purified EGFP-MAD1 interacted with His-HA-RED. Affinity-purified EGFP (G) or EGFP-MAD1 (M) fusion proteins were incubated with purified His-HA-RED recombinant proteins (see “Experimental Procedures”). The precipitated proteins were analyzed by immunoblot. Purified EGFP, EGFP-MAD1, and His-HA-RED recombinant proteins were loaded as input controls. C and D, MAD1 binds to RED in vivo. Nocodazole-arrested mitotic cells were shaken off and released from arrest in fresh medium lacking nocodazole for ∼40 min. Mitotic cells were collected again for coimmunoprecipitation assays and DNA content analysis by flow cytometry. About 2 mg of cell lysates were used for immunoprecipitation (IP) assay. Immunoprecipitated proteins were determined by immunoblot. C, RED was coimmunoprecipitated with the purified rabbit anti-MAD1 antibodies using protein G beads from mitotic cell lysates. The purified rabbit non-immune (non-I) antibody was used as a control. Eighty micrograms of cell lysates were loaded as an input control. The asterisks indicate the light chain of rabbit IgG. D, MAD1 was coimmunoprecipitated with the purified rabbit anti-RED antibodies using protein G beads from mitotic cell lysates. 45 μg of cell lysates was loaded as an input control.
Next, using rabbit anti-MAD1 and anti-RED antibodies, we performed coimmunoprecipitation assays to determine whether MAD1 can bind to RED in vivo. Considering the observation that RED and MAD1 colocalized to the spindle poles in metaphase and anaphase (see Fig. 4A), cells were arrested in prometaphase by nocodazole and subsequently released from prometaphase arrest for collection of mitotic cells at stages later than prometaphase for coimmunoprecipitation assays and flow cytometric analyses. We found that RED coprecipitated with MAD1 from the mitotic cell lysates, as did MAD2, when rabbit anti-MAD1 antibodies were used (Fig. 2C), and MAD1 coprecipitated with RED when rabbit anti-RED antibodies were used (Fig. 2D). The DNA content of the mitotic cells was also confirmed by flow cytometric analysis (Fig. 2, C and D). In addition, we also found that MAD1 did not interact with RED in either the early S-phase or in prometaphase cells, whereas MAD1 interacted with MAD2 (see supplemental Fig. S2). These results indicate that MAD1 interacts with RED in mitotic cells at the mitotic stages later than prometaphase. Taken together, our results demonstrate that MAD1 interacts with RED both in vivo and in vitro.
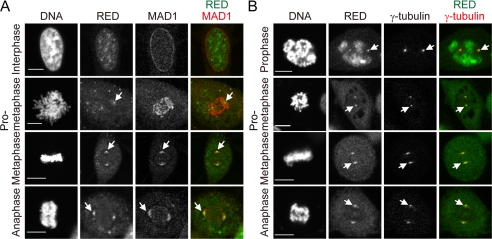
FIGURE 4.
Intracellular localization of RED. A, colocalization of MAD1 and RED at the spindle poles in metaphase and anaphase. Cells were fixed with paraformaldehyde and simultaneously stained with rat anti-MAD1 and rabbit anti-RED antibodies, followed by Cy3-conjugated anti-rat IgG and FITC-conjugated anti-rabbit IgG antibodies. Chromosomal DNA was stained with DAPI. Stained cells were visualized by confocal laser microscope. The arrows indicate the locations of the spindle poles. Scale bar = 10 μm. B, localization of RED at the spindle poles. Cells were fixed with cold methanol and simultaneously stained with rabbit anti-RED and mouse anti-γ-tubulin antibodies, followed by FITC-conjugated anti-rabbit IgG and Cy3-conjugated anti-mouse IgG antibodies. Arrows indicate the locations of the spindle poles. Scale bar = 10 μm.
The RED-binding Domain in MAD1
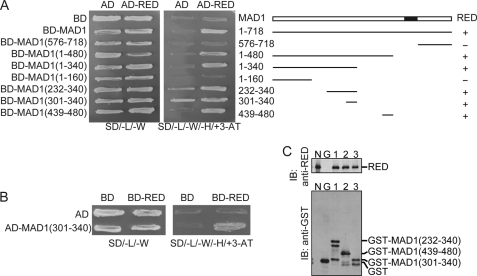
To determine the RED-binding domain in MAD1, a series of MAD1 truncation proteins were fused to the C terminus of the Gal4BD or Gal4AD for yeast two-hybrid analyses (Fig. 3, A and B). It was initially found that a small region from amino acid residues 439–480 of MAD1, which was designated as MAD1(439–480), could interact with RED (Fig. 3A). However, it was also observed that BD-MAD1(301–340) or BD-MAD1(232–340), itself, could activate expression of the HIS3 reporter gene in yeast (Fig. 3A). MAD1(301–340) was subsequently fused to the Gal4 activation domain, and RED was fused to the Gal4 DNA-binding domain for a yeast two-hybrid analysis (Fig. 3B). We found that MAD1(301–340) did, indeed, interact with RED (Fig. 3B).
FIGURE 3.
Yeast two-hybrid analysis and GST pull-down assays for the identification of RED-interacting regions in MAD1. A, a schematic map of MAD1 and its truncation alleles used to define the RED-interacting region in yeast two-hybrid analysis. Full-length MAD1 was fused to the DNA- BD of the yeast Gal4 transcription factor and RED to the AD of yeast Gal4. A series of MAD1 truncation proteins were fused to the DNA-binding domain of the Gal4 transcription factor for determination of the RED-interacting region of MAD1. Synthetic dextrose minimal solid medium (SD) lacking leucine and tryptophan (SD/-L/-W) was used to cultivate yeast transformants, and SD medium lacking leucine, tryptophan, and histidine and containing 25 mm 3-aminotriazole (SD/-L/-W/-H/+3AT) was used to assay for expression of a HIS3 reporter gene. +, interaction between various MAD1 truncation proteins and RED; -, no interaction. The closed box indicates the MAD2-binding region. B, confirmation of the interaction between MAD1(301–340) and RED. Both BD-RED and AD-MAD1(301–340) vectors were used for yeast two-hybrid analysis. C, MAD1(301–340) and MAD1(439–480) bind to RED in vitro. Purified GST (G), GST-MAD1(232–340) (1), GST-MAD1(439–480) (2), and GST-MAD1(301–340) (3) were incubated with purified His-HA-RED recombinant protein in RIPA buffer and precipitated with glutathione-conjugated Sepharose beads at 4 °C. The precipitated proteins were analyzed by immunoblot (IB). Purified His-HA-RED recombinant protein was loaded as an input control (N).
To further confirm that MAD1(232–340), MAD1(301–340), and MAD1(439–480) interacted with RED, the GST-MAD1(232–340), GST-MAD1(301–340), and GST-MAD1(439–480) fusion proteins were overexpressed in and purified from bacteria for in vitro binding analyses. Consistently, the GST-MAD1(232–340), GST-MAD1(301–340), and GST-MAD1(439–480) fusion proteins were found to interact with the RED recombinant protein in vitro, whereas the GST protein could not (Fig. 3C).
Subcellular Localization of RED
To determine whether MAD1 colocalized with RED during cell cycle progression, the rat anti-MAD1 and rabbit anti-RED antibodies were simultaneously used to stain MAD1 and RED, respectively, in HeLa cells (Fig. 4A). It was found that MAD1 localized to the nuclear envelope, whereas RED was observed within the nucleus during interphase. In prometaphase, MAD1 localized to kinetochores, whereas RED appeared to be at the spindle poles and in the cytoplasm. Thus, MAD1 and RED did not colocalize in the nucleus during interphase or to kinetochores in prometaphase. Nevertheless, MAD1 and RED appeared to colocalize to the spindle poles in metaphase and anaphase.
To confirm that RED localized to the spindle poles during mitosis, mouse anti-γ-tubulin and rabbit anti-RED antibodies were simultaneously used to stain γ-tubulin, a component of the spindle poles, and RED, respectively. We found that RED colocalized with γ-tubulin at the spindle poles from prophase to anaphase (Fig. 4B).
To further verify that RED localizes to the spindle poles but not to the kinetochore in prometaphase, we examined the subcellular localization of ectopically expressed EGFP-RED in mitotic cells. Consistently, EGFP-RED did not associate with kinetochores but did localize to the spindle poles in prometaphase (supplemental Fig. S3). Therefore, we concluded that RED is a spindle pole-associated protein but not a kinetochore-associated protein.
RED-silenced Cells May Bypass the Mitotic Arrest Induced by Nocodazole
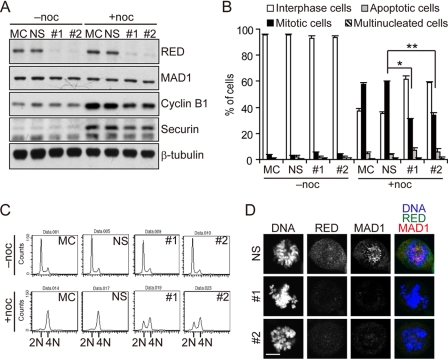
To examine the role of RED in the SAC, two RED siRNA duplexes (#1 and #2, see “Experimental Procedures”) were used to deplete the levels of the RED protein. Depletion of RED did not affect the protein levels of MAD1 (Fig. 5A) and did not result in cell cycle arrest or cell death after cells were treated with RED siRNA duplexes for 42 h (Fig. 5B). Similarly, a flow cytometric analysis revealed that the DNA content histogram profiles were not appreciably different between the non-silenced and RED-depleted cells (Fig. 5C). However, cells went to apoptosis after treatment with RED siRNA duplexes for 3 days (supplemental Fig. S4). Thus, to determine the role of RED in the SAC, we performed the experiments with RED siRNA duplexes within 2 days (see below).
FIGURE 5.
Effect of RED RNAi on the SAC. HeLa cells were mock-treated (MC) or transfected with non-silencing (NS) siRNA or RED siRNA#1 or #2 for 24 h before treatment with (+) or without (-) nocodazole. For each transfection, 200 pmols of the siRNA duplexes were used. Cells were collected for analysis after treatment with or without nocodazole (noc) for 18 h. A, depletion of RED protein. Cells were lysed and analyzed by Western blotting with antibodies against RED, MAD1, cyclin B1, securin, and β-tubulin (loading control). B, quantification of mitotic cells in mock-, non-silencing siRNA-, RED siRNA#1-, or RED siRNA#2-treated cell cultures. Two hundred cells were scored for each of three independent experiments. Data shown in graph are the mean ± S.D. from three independent experiments. *, p < 0.001; **, p < 0.001 (analysis of variance). C, DNA-content histograms for mock-, non-silencing siRNA-, RED siRNA#1-, or RED siRNA#2-treated cells analyzed by flow cytometry. D, localization of MAD1 in RED-depleted prometaphase cells. Staining for MAD1 and RED was performed as described in Fig. 4A. Scale bar = 10 μm.
We next asked whether the RED-depleted cells failed to arrest in mitosis after treatment with the microtubule destabilizer nocodazole, which is a phenotype of SAC-defective cells (19, 24–31). After an 18-h treatment with nocodazole, ∼31% of the RED siRNA#1-treated and ∼34% of RED siRNA#2-treated cells were observed to be in mitosis, whereas ∼58% of the mock-treated cells and ∼60% of the non-silencing siRNA-treated cells were in mitosis (Fig. 5B). Furthermore, ∼62% of the RED siRNA#1-treated and ∼59% of the RED siRNA#2-treated cells were in interphase, compared with ∼37% of the mock-treated cells and ∼36% of the non-silencing siRNA-treated cells (Fig. 5B). Flow cytometric analysis also showed that after nocodazole treatment, ∼34% of the RED siRNA#1-treated and ∼35% of the RED siRNA#2-treated cells were in G1, whereas ∼6% of the mock-treated and ∼9% of the non-silencing siRNA-treated cells were in G1 (Fig. 5C). In addition, the protein levels of both cyclin B1 and securin were lower in the RED-depleted cells than in the mock-treated or non-silencing siRNA-treated cells after nocodazole treatment (Fig. 5A). Therefore, we concluded that compared with the mock-treated or non-silencing siRNA-treated cells, the SAC was defective in the RED-depleted cells, and, consequently, the anaphase-promoting complex/cyclosome can promote the proteolysis of securin and cyclin B1 to cause cells to bypass the mitotic arrest induced by nocodazole and complete mitosis for entry into the next round of the cell cycle. Additionally, the defect in the SAC caused by the siRNA#1-directed depletion of RED could be rescued by an siRNA#1-resistant RED construct (see “Experimental Procedures” and supplemental Fig. S5). This finding indicates that the defective SAC caused by the siRNA#1-directed depletion of RED was not due to an off-target inhibition. We also determined the response of the RED-depleted cells to different amounts of nocodazole and found that the decreased percentage of mitotic cells was due to a defect in the SAC caused by the depletion of RED but not due to the effect of RED depletion on nocodazole sensitivity (see Supplemental Fig. S6). Taken together, these results suggest that RED is required for activation of the SAC in prometaphase.
By immunofluorescence microscopy, we found that MAD1 failed to localize to kinetochores in ∼81% (39 of 48) of the RED siRNA#1-treated and ∼87% (39 of 45) of the RED siRNA#2-treated prometaphase cells, whereas RED normally localized to the spindle poles in the MAD1-depleted prometaphase cells (Fig. 5D and supplemental Figs. S7 and S8A). Similarly, MAD1 could not normally localize to kinetochores in ∼78% (21 of 27) of the nocodazole-treated RED-depleted prometaphase cells (supplemental Fig. S8B). Consistent with the previous observation that MAD1 is required to recruit MAD2 to kinetochores (13), we found that MAD2 also failed to localize to kinetochores in ∼92% (48 of 52) of the RED-depleted prometaphase cells (supplemental Fig. S9). These data indicate that RED determines the kinetochore localization of MAD1 and MAD2, whereas the localization of RED to the spindle poles is independent of MAD1. This outcome implies that depletion of RED may interfere with the ability of MAD1 and, thereby, of MAD2 to properly localize to kinetochores and thus causes a defect in the SAC to allow cells to bypass a nocodazole-induced mitotic arrest. In addition, MAD1 failed to properly localize to the spindle poles in ∼74% (20 of 27) of the RED-depleted metaphase cells (supplemental Fig. S10), indicating that depletion of RED might cause a failure in the localization of MAD1 to the spindle poles in metaphase cells.
Silencing of RED by RNA Interference Results in a Change in Mitosis Progression
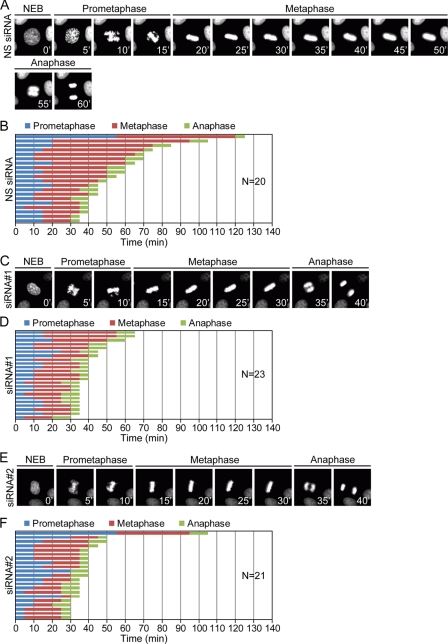
To determine whether depletion of RED could affect mitosis progression, we measured the timing from the nuclear envelope breakdown (NEBD) to anaphase in both the non-silenced and RED-depleted cells. After cells were treated with the siRNA duplexes for 24 h and thereafter arrested in the early S-phase by treatment with thymidine, cells were released from the early S-phase to measure the timing from the NEBD to anaphase using a live time-lapse fluorescence microscope. We found that the average time from NEBD to anaphase onset in the non-silenced cells was ∼52.5 min (prometaphase, ∼16.3 min; metaphase, ∼36.2 min; n = 20), whereas the average time from the NEBD to anaphase onset in the siRNA#1-treated cells and the siRNA#2-treated cells was ∼34.1 min (prometaphase, ∼12.4 min; metaphase, ∼21.7 min; n = 23) and ∼33.8 min (prometaphase, ∼15.7 min; metaphase, ∼18.1 min; n = 21), respectively (Fig. 6). The timing of both prometaphase and metaphase in the RED-depleted cells was shorter compared with the non-silenced cells, indicating that depletion of RED could shorten the timing from the NEBD to anaphase onset.
FIGURE 6.
Effect of RED RNAi on mitotic progression. H2B-GFP HeLa cells were transfected with the indicated siRNA duplexes and then arrested in early S-phase by thymidine. Cells were released from early S-phase arrest for measurement of mitotic timing using live cell time-lapse fluorescence microscopy. NEBD was designated as T0, and the timing from NEBD to anaphase was measured from live cell movies. A, C, and E, successive frames every 5 min from live cell videos of H2B-GFP HeLa cells transfected with indicated siRNA duplexes. B, D, and F, mitotic progression of H2B-GFP HeLa cells transfected with indicated siRNA duplexes. Timing for each group of cells, selected randomly, is shown in graphs. N, number of cells selected at random from two independent experiments.
We also determined the doubling time of the RED-depleted cells by a live cell imaging system. We found that the doubling time of non-silenced cells, siRNA#1-treated cells, and siRNA#2-treated cells was ∼942, ∼921, and ∼938 min, respectively (supplemental Fig. S11). This indicates that depletion of RED does not affect progression of cell cycle phases other than the M phase.
Many SAC proteins are known to be required for chromosome alignment, and their depletion by RNAi may result in chromosome misalignment in metaphase, thereby resulting in lagging chromosomes in anaphase (25–27, 30, 32–34). However, in our live cell images (Fig. 6), the RED-depleted metaphase cells could align chromosomes normally before proceeding into anaphase, and the RED-depleted anaphase cells did not have lagging chromosome(s). This indicates that RED is not required for chromosome alignment and proper chromosome segregation.
MAD1(301–340) or MAD1(439–480) Peptides Fused to EGFP Are Capable of Binding to RED and Cause Cells to Bypass the Mitotic Arrest Induced by Nocodazole
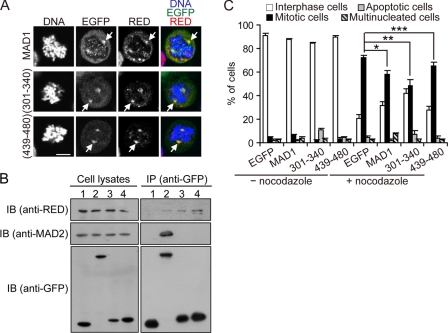
We found that moderate expression or overexpression of MAD1, which was fused to the C terminus of EGFP, could cause a defect in the SAC (supplemental Fig. S12). Additionally, overexpression of MAD1 caused a greater defect in the SAC than moderate expression of MAD1 (supplemental Fig. S12). Importantly, ectopically expressed EGFP-MAD1 was able to colocalize with RED at the spindle poles in prometaphase (Fig. 7A). This result raised the possibility that overexpression of EGFP-MAD1 caused a SAC defect by inappropriately inactivating RED at the spindle poles. Because several MAD1-interacting proteins have been observed previously as well as in this study (12, 13, 35), we tested this possibility by determining whether ectopic expression of the RED-interacting peptide MAD1(301–340) or MAD1(439–480) fused to EGFP could cause a SAC defect. To avoid the severe lethality caused by overexpression of EGFP-MAD1(301–340) (data not shown), which could confound our analysis, translation of EGFP or its fusion genes was directed by the partially disabled IRES of the encephalomyocarditis virus to yield moderate protein expression levels (18). Notably, moderate expression of EGFP-MAD1(301–340) caused only a mild lethality and did not result in a significant change in the mitotic population (Fig. 7C).
FIGURE 7.
Effect of ectopic expression of MAD1 and its derivatives on the role of RED in the SAC. Cells were transfected with IRES-EGFP (1), IRES-EGFP-MAD1 (2), IRES-EGFP-MAD1(301–340) (3) or IRES-EGFP-MAD1(439–480) (4). For each transfection, 300 ng of plasmid DNA were used. A, intracellular localization of EGFP-MAD1, EGFP-MAD1(301–340), and EGFP-MAD1(439–480). Cells were stained for DNA, EGFP fusion proteins, and RED. Arrows indicate the spindle poles. Scale bar, 10 μm. B, association of RED with EGFP-MAD1(301–340) and EGFP-MAD1(439–480). Cells were lysed for coimmunoprecipitation assays using GFP-Trap®_A beads. The precipitated proteins were analyzed by immunoblot (IB). C, effect of moderate expression of EGFP-MAD1(301–340) and EGFP-MAD1(439–480) on the SAC. 24 h after transfection, cells were treated with dimethyl sulfoxide or nocodazole for 18 h and then collected for analysis. Two hundred GFP-positive cells were scored for each of three independent experiments. Data shown in the graph are the mean ± S.D. from three independent experiments. *, p < 0.001; **, p < 0.001; ***, p = 0.038 (analysis of variance).
We initially examined the localization of EGFP-MAD1(301–340) and EGFP-MAD1(439–480) in prometaphase and found that both colocalized with RED at the spindle poles (Fig. 7A). Additionally, neither EGFP-MAD1(301–340) nor EGFP-MAD1(439–480) colocalized with endogenous MAD1 proteins at kinetochores or interfered with the kinetochore localization of MAD1 (supplemental Fig. S13). We then performed coimmunoprecipitation assays using anti-GFP antibody-conjugated beads to determine whether EGFP-MAD1(301–340) and EGFP-MAD1(439–480) could bind to RED or MAD2 in cells. Consistent with the results from the in vitro binding assays (Fig. 3C), EGFP-MAD1(301–340) and EGFP-MAD1(439–480) were able to interact with RED in vivo, as could EGFP-MAD1 (Fig. 7B). Unlike EGFP-MAD1, EGFP-MAD1(301–340) and EGFP-MAD1(439–480) did not associate with MAD2 (Fig. 7B).
We then determined whether moderate expression of EGFP-MAD1(301–340) or EGFP-MAD1(439–480) could cause cells to fail to arrest in mitosis after treatment with nocodazole. After 18 h of nocodazole treatment, ∼72% of the cells expressing EGFP, ∼58% of the cells expressing EGFP-MAD1, ∼48% of the cells expressing EGFP-MAD1(301–340) and ∼65% of the cells expressing EGFP-MAD1(439–480) were in mitosis (Fig. 7C), whereas∼21% of the cells expressing EGFP, ∼32% of the cells expressing EGFP-MAD1, ∼42% of the cells expressing EGFP-MAD1(301–340) and ∼28% of the cells expressing EGFP-MAD1(439–480) were in interphase (C). Therefore, we conclude that compared with EGFP, moderate expression of EGFP-MAD1, EGFP-MAD1(301–340) or EGFP-MAD1(439–480) may cause a defect in the SAC.
DISCUSSION
In this report, we showed that the nuclear protein RED interacted with the mitotic checkpoint protein MAD1, and its expression was cell cycle-regulated (Figs. 1 and 2). RED was not found to be a kinetochore-associated protein, and it localized to the spindle poles and cytoplasm during mitosis (Fig. 4 and supplemental Fig. S3). RED and MAD1 only colocalized at the spindle poles in metaphase and anaphase (Fig. 4). We found that there are two small regions within MAD1, MAD1(301–340) and MAD1(439–480) that are capable of binding to RED (Figs. 3 and 7B). Furthermore, these two small RED-interacting regions do not overlap with the two leucine zipper motifs (501–520 and 557–571) that are required for MAD2 binding to MAD1 (36). Our findings also confirmed that MAD1(301–340) and MAD1(439–480) do not associate with MAD2 in vivo (Fig. 7B).
Although RED is a nonkinetochore protein, two lines of evidence suggests that RED has a functional role in the activation of the SAC. First, we have shown that EGFP-MAD1(301–340) and EGFP-MAD1(439–480) can associate with RED at the spindle poles in prometaphase and, hence, abrogate the SAC (Fig. 7, A–C). Furthermore, neither EGFP-MAD1(301–340) nor EGFP-MAD1(439–480) colocalized with endogenous MAD1 at the kinetochores or interfered with the kinetochore localization of MAD1 in prometaphase (supplemental Fig. S13). Additionally, depletion of MAD1 did not interfere with the localization of RED to the spindle poles in prometaphase (supplemental Fig. S7). Therefore, it is possible that the spindle pole-associated portion of RED proteins may execute the SAC. Second, depletion of RED may cause a failure in the kinetochore localization of MAD1 and MAD2 (Fig. 5D and supplemental Figs. S8 and S9) and a defect in the SAC (Fig. 5, B and C), suggesting that RED may determine the kinetochore localization of MAD1 and MAD2 for the activation of the SAC at the kinetochores in prometaphase. Nevertheless, because RED proteins are in the cytosol and at the spindle poles in prometaphase (Fig. 4), it remains to be determined whether either a cytosolic or spindle pole-associated portion of RED proteins is required for the recruitment of MAD1 to kinetochores.
At present, we do not know how RED determines the kinetochore localization of MAD1. Our observation that RED does not interact with MAD1 from interphase to prometaphase indicates that the recruitment of MAD1 to kinetochores does not depend upon its physical interaction with RED (Fig. 4A and supplemental Fig. S2). Previous studies have shown that during the kinetochore assembly cascade in prometaphase, recruitment of MAD1 to the kinetochores is determined by CENP-F, the NDC80/HEC1 complex, and the Rod-Zw10-Zwilch complex, which localize to the kinetochores (13, 22, 37–39). Because RED is a nonkinetochore protein (Fig. 4A), it is unlikely that RED directly regulates CENP-F, the NDC80/HEC1 complex, or the Rod-Zw10-Zwilch complex to determine the kinetochore localization of MAD1. However, RED has been identified to interact with the nuclear pore protein NUP62, which may bind to NUP358/RanBP2, a kinetochore protein that is required for the kinetochore localization of the SAC proteins MAD1, MAD2, and ZW10 (40, 41), and to the mitotic checkpoint regulator Rae1, which may interact with BUB1, a checkpoint protein required for the kinetochore localization of MAD1 and MAD2 (34, 42–45). Therefore, it is possible that RED determines the kinetochore localization of MAD1 via the NUP62-RanBP2 and/or NUP62-Rae1 interactions. Nevertheless, the role of NUP62 in the SAC or chromosome alignment remains to be uncovered.
Several SAC proteins (e.g. MAD2 and BUBR1) have been shown to play a role in controlling the mitotic timing in addition to their roles in the SAC (25, 28). Furthermore, it appears that cytosolic, but not kinetochore-associated, MAD2 and BUBR1 regulate mitotic timing (28). A recent study demonstrates that in Drosophila, the spindle, SAC, and mitotic timer functions of BubR1 are separable (46). Here, we also find that RED has a role in controlling mitotic timing, as judged by our observation that depletion of RED accelerates mitosis progression from the NEBD to anaphase (Fig. 6). Because RED proteins are present at the spindle poles and in the cytoplasm in mitotic cells (Fig. 4), it is also possible that, similar to MAD2 and BUBR1 (25, 28), the SAC and timer functions of RED are separable. It remains to be determined whether cytosolic RED can regulate the timing from NEBD to anaphase onset as cytosolic MAD2 and BUBR1 can.
Many known SAC proteins (e.g. MAD1 and BUB1) are required for chromosome alignment and segregation (25–27, 30, 32–34). Nevertheless, it is puzzling that the RED-depleted metaphase cells can align chromosomes normally before proceeding into anaphase and that the RED-depleted anaphase cells do not have lagging chromosome(s) despite the failure of MAD1 and MAD2 to localize to kinetochores and the defect in the SAC (Figs. 5–6 and supplemental Figs. S8 and S9). We speculate that MAD1 should also have two separable functions: one for activation of the SAC in a kinetochore-dependent manner and the other for chromosome alignment and segregation in a kinetochore-independent manner. This speculation is on the basis of two previous observations. One observation has shown that the bubR1-KEN mad2 double mutant flies can accurately segregate chromosomes although they lack the SAC function (46). The other observation has revealed that in human BUB1, deletion of its conserved domain I (amino acids 458–476) nearly abrogates the SAC function but can efficiently regulate chromosome alignment (34). These two observations indicate that the defective SAC does not necessarily affect chromosome alignment and segregation and may explain why the RED-depleted cells can execute chromosome alignment and segregation normally even though MAD1 and MAD2 fail to localize to the kinetochores and the SAC is defective.
It is often observed that the SAC-defective cells may go to apoptosis or form multinuclei when treated with nocodazole. However, this phenotype was not observed in the RED-depleted cells. Thus, as a comparison, we simultaneously determined the SAC-defectiveness of both MAD1- and RED-depleted cells (supplemental Fig. S14). In our analysis, depletion of MAD1 induced by the MAD1 siRNA used in this study did not result in a more severe defect in the SAC than depletion of RED (supplemental Fig. S14). Similarly, the MAD1-depleted cells also did not go to apoptosis or form multinuclei when treated with nocodazole (supplemental Fig. S14). It should be noted that the RED- or MAD1-depleted cells did not have a severe defect in the SAC in this study, which is unlike the HCT-MAD2+/− or BUB1-depleted cells (34, 47). Additionally, one study has shown that depletion of HEC1 has a more severe defect in the SAC than that of Zwint-1, and depletion of HEC1 may induce formation of more multinuclei than that of Zwint-1 after treatment with nocodazole (26). In addition, another previous study has shown that depletion of BUB1 may cause a severe defect in the SAC and induce apoptosis and that expression of the BUB1A130S and BUB1Y259C mutant proteins may cause a mild defect in the SAC but does not induce apoptosis in human cells (34). Therefore, a mild defect in the SAC caused by depletion of RED may not induce apoptosis or formation of multinuclei upon nocodazole treatment.
It is known that MAD1 is released from the spindle-attached kinetochores and relocalized to the spindle poles for silencing of the SAC in metaphase (1, 4). However, it is not clear why MAD1 has to be relocalized to the spindle poles in metaphase. Our observation that MAD1 interacted with RED at the spindle poles in metaphase (Fig. 4) implies that RED should be at least one of the targets of MAD1 at the spindle poles. Previous observations have shown that in Xenopus egg extracts, the observed defect in the SAC, which was caused by excessive amounts of MAD1, was due to its titrating out free MAD2 from the cytosol (48). Similarly, overexpression of EGFP-MAD1 also caused a considerable defect in the SAC (supplemental Fig. S12), and, surprisingly, unlike endogenous MAD1 (see Fig. 4A and Ref. 20), EGFP-MAD1 was able to not only localize to kinetochores but also to the spindle poles in prometaphase (Fig. 7). Furthermore, EGFP-MAD1(301–340) and EGFP-MAD1(439–480) were observed to bind to and colocalize with RED at the spindle poles in prometaphase, and their expression could result in a defect in the SAC (Figs. 3 and 7). Therefore, it is possible that, in addition to titrating out free MAD2 from the cytosol, the localization of EGFP-MAD1 to the spindle poles could also be attributed to the defect in the SAC caused by overexpression of EGFP-MAD1, which was likely due to its interaction with RED at the spindle poles in prometaphase. In addition, depletion of RED could cause a failure in localization of MAD1 to the spindle poles in metaphase (see supplemental Fig. S10). Taken together, these observations also lead to speculation that MAD1 may bind to and inactivate RED to silence the SAC when MAD1 is relocalized to the spindle poles in metaphase. This speculation warrants further investigation in the future.
The concept that the core and additional components of the SAC are recruited to kinetochores to ensure a proper attachment of the paired sister chromatids to microtubules during prometaphase is well known (1). However, a previous observation revealed that Death inducer-obliterator polypeptide 3 (Dido3), a nonkinetochore protein, has been shown to be part of the network of regulatory molecules involved in activation of the SAC at the spindle poles (49). Like Dido3, RED also has a role in the SAC at the spindle poles during prometaphase (Fig. 7). More importantly, our finding that RED is required for the localization of MAD1 to kinetochores even though they are at different locations and do not interact with each other in prometaphase (Fig. 5D and supplemental Fig. S2) suggests that a RED-dependent signaling pathway, possibly via NUP62 (see above mentioned description), from a non-kinetochore location to the kinetochore should exist for activation of the SAC. Taken together, these observations reveal that non-kinetochore proteins must coordinate with kinetochore proteins for the execution of the SAC in prometaphase.
Supplementary Material
Acknowledgments
We thank Drs. T. Yen and D. Pellman for providing the plasmids and Dr. G. Wahl for providing H2B-GFP HeLa cells.
This work was supported by grants from the National Health Research Institute, Taiwan (NHRI-EX95–9101BC) and the Tzu-Chi Foundation (to Y. L. J.).

This article contains supplemental Figs. S1–S14.
- SAC
- spindle assembly checkpoint
- BD
- binding domain
- AD
- activation domain
- aa
- amino acids
- IRES
- internal ribosome entry sequence
- RIPA
- radioimmune precipitation assay
- NEBD
- nuclear envelope breakdown.
REFERENCES
- 1. Musacchio A., Salmon E. D. (2007) The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8, 379–393 [DOI] [PubMed] [Google Scholar]
- 2. Howell B. J., McEwen B. F., Canman J. C., Hoffman D. B., Farrar E. M., Rieder C. L., Salmon E. D. (2001) Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol. 155, 1159–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Howell B. J., Moree B., Farrar E. M., Stewart S., Fang G., Salmon E. D. (2004) Spindle checkpoint protein dynamics at kinetochores in living cells. Curr. Biol. 14, 953–964 [DOI] [PubMed] [Google Scholar]
- 4. Shah J. V., Botvinick E., Bonday Z., Furnari F., Berns M., Cleveland D. W. (2004) Dynamics of centromere and kinetochore proteins. Implications for checkpoint signaling and silencing. Curr. Biol. 14, 942–952 [DOI] [PubMed] [Google Scholar]
- 5. Habu T., Kim S. H., Weinstein J., Matsumoto T. (2002) Identification of a MAD2-binding protein, CMT2, and its role in mitosis. EMBO J. 21, 6419–6428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mapelli M., Filipp F. V., Rancati G., Massimiliano L., Nezi L., Stier G., Hagan R. S., Confalonieri S., Piatti S., Sattler M., Musacchio A. (2006) Determinants of conformational dimerization of Mad2 and its inhibition by p31comet. EMBO J. 25, 1273–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xia G., Luo X., Habu T., Rizo J., Matsumoto T., Yu H. (2004) Conformation-specific binding of p31(comet) antagonizes the function of Mad2 in the spindle checkpoint. EMBO J. 23, 3133–3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yang M., Li B., Tomchick D. R., Machius M., Rizo J., Yu H., Luo X. (2007) p31comet blocks Mad2 activation through structural mimicry. Cell 131, 744–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hagting A., Den Elzen N., Vodermaier H. C., Waizenegger I. C., Peters J. M., Pines J. (2002) Human securin proteolysis is controlled by the spindle checkpoint and reveals when the APC/C switches from activation by Cdc20 to Cdh1. J. Cell Biol. 157, 1125–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Waizenegger I. C., Hauf S., Meinke A., Peters J. M. (2000) Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell 103, 399–410 [DOI] [PubMed] [Google Scholar]
- 11. Hauf S., Waizenegger I. C., Peters J. M. (2001) Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science 293, 1320–1323 [DOI] [PubMed] [Google Scholar]
- 12. Jin D. Y., Spencer F., Jeang K. T. (1998) Human T cell leukemia virus type 1 oncoprotein Tax targets the human mitotic checkpoint protein MAD1. Cell 93, 81–91 [DOI] [PubMed] [Google Scholar]
- 13. Martin-Lluesma S., Stucke V. M., Nigg E. A. (2002) Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science 297, 2267–2270 [DOI] [PubMed] [Google Scholar]
- 14. Assier E., Bouzinba-Segard H., Stolzenberg M. C., Stephens R., Bardos J., Freemont P., Charron D., Trowsdale J., Rich T. (1999) Isolation, sequencing and expression of RED, a novel human gene encoding an acidic-basic dipeptide repeat. Gene 230, 145–154 [DOI] [PubMed] [Google Scholar]
- 15. Krief P., Augery-Bourget Y., Plaisance S., Merck M. F., Assier E., Tanchou V., Billard M., Boucheix C., Jasmin C., Azzarone B. (1994) A new cytokine (IK) down-regulating HLA class II: monoclonal antibodies, cloning and chromosome localization. Oncogene 9, 3449–3456 [PubMed] [Google Scholar]
- 16. Neubauer G., King A., Rappsilber J., Calvio C., Watson M., Ajuh P., Sleeman J., Lamond A., Mann M. (1998) Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nat. Genet. 20, 46–50 [DOI] [PubMed] [Google Scholar]
- 17. Zhou Z., Licklider L. J., Gygi S. P., Reed R. (2002) Comprehensive proteomic analysis of the human spliceosome. Nature 419, 182–185 [DOI] [PubMed] [Google Scholar]
- 18. Rees S., Coote J., Stables J., Goodson S., Harris S., Lee M. G. (1996) Bicistronic vector for the creation of stable mammalian cell lines that predisposes all antibiotic-resistant cells to express recombinant protein. BioTechniques 20, 102–110 [DOI] [PubMed] [Google Scholar]
- 19. Luo X., Tang Z., Rizo J., Yu H. (2002) The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol. Cell 9, 59–71 [DOI] [PubMed] [Google Scholar]
- 20. Campbell M. S., Chan G. K., Yen T. J. (2001) Mitotic checkpoint proteins HsMAD1 and HsMAD2 are associated with nuclear pore complexes in interphase. J. Cell Sci. 114, 953–963 [DOI] [PubMed] [Google Scholar]
- 21. Rogers L. (1998) in Cells: A Laboratory Manual (Spector D. L., Dolgman R. D., Leineand L. A., eds.) pp 16.11–16.12, Cold Spring Harbor Laboratory Press, Plainview, New York [Google Scholar]
- 22. Laoukili J., Kooistra M. R., Brás A., Kauw J., Kerkhoven R. M., Morrison A., Clevers H., Medema R. H. (2005) FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat. Cell Biol. 7, 126–136 [DOI] [PubMed] [Google Scholar]
- 23. Mayor T., Stierhof Y. D., Tanaka K., Fry A. M., Nigg E. A. (2000) The centrosomal protein C-Nap1 is required for cell cycle-regulated centrosome cohesion. J. Cell Biol. 151, 837–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chan G. K., Jablonski S. A., Starr D. A., Goldberg M. L., Yen T. J. (2000) Human Zw10 and ROD are mitotic checkpoint proteins that bind to kinetochores. Nat. Cell Biol. 2, 944–947 [DOI] [PubMed] [Google Scholar]
- 25. Kiyomitsu T., Obuse C., Yanagida M. (2007) Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev. Cell 13, 663–676 [DOI] [PubMed] [Google Scholar]
- 26. Lin Y. T., Chen Y., Wu G., Lee W. H. (2006) Hec1 sequentially recruits Zwint-1 and ZW10 to kinetochores for faithful chromosome segregation and spindle checkpoint control. Oncogene 25, 6901–6914 [DOI] [PubMed] [Google Scholar]
- 27. McAinsh A. D., Meraldi P., Draviam V. M., Toso A., Sorger P. K. (2006) The human kinetochore proteins Nnf1R and Mcm21R are required for accurate chromosome segregation. EMBO J. 25, 4033–4049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Meraldi P., Draviam V. M., Sorger P. K. (2004) Timing and checkpoints in the regulation of mitotic progression. Dev. Cell 7, 45–60 [DOI] [PubMed] [Google Scholar]
- 29. Montembault E., Dutertre S., Prigent C., Giet R. (2007) PRP4 is a spindle assembly checkpoint protein required for MPS1, MAD1, and MAD2 localization to the kinetochores. J. Cell Biol. 179, 601–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Osmundson E. C., Ray D., Moore F. E., Gao Q., Thomsen G. H., Kiyokawa H. (2008) The HECT E3 ligase Smurf2 is required for Mad2-dependent spindle assembly checkpoint. J. Cell Biol. 183, 267–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stucke V. M., Silljé H. H., Arnaud L., Nigg E. A. (2002) Human Mps1 kinase is required for the spindle assembly checkpoint but not for centrosome duplication. EMBO J. 21, 1723–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee S. H., Sterling H., Burlingame A., McCormick F. (2008) Tpr directly binds to Mad1 and Mad2 and is important for the Mad1-Mad2-mediated mitotic spindle checkpoint. Genes Dev. 22, 2926–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Logarinho E., Resende T., Torres C., Bousbaa H. (2008) The human spindle assembly checkpoint protein Bub3 is required for the establishment of efficient kinetochore-microtubule attachments. Mol. Biol. Cell 19, 1798–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Klebig C., Korinth D., Meraldi P. (2009) Bub1 regulates chromosome segregation in a kinetochore-independent manner. J. Cell Biol. 185, 841–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lou Y., Yao J., Zereshki A., Dou Z., Ahmed K., Wang H., Hu J., Wang Y., Yao X. (2004) NEK2A interacts with MAD1 and possibly functions as a novel integrator of the spindle checkpoint signaling. J. Biol. Chem. 279, 20049–20057 [DOI] [PubMed] [Google Scholar]
- 36. Iwanaga Y., Kasai T., Kibler K., Jeang K. T. (2002) Characterization of regions in hsMAD1 needed for binding hsMAD2. A polymorphic change in an hsMAD1 leucine zipper affects MAD1-MAD2 interaction and spindle checkpoint function. J. Biol. Chem. 277, 31005–31013 [DOI] [PubMed] [Google Scholar]
- 37. Chen T. C., Lee S. A., Chan C. H., Juang Y. L., Hong Y. R., Huang Y. H., Lai J. M., Kao C. Y., Huang C. Y. (2009) Cliques in mitotic spindle network bring kinetochore-associated complexes to form dependence pathway. Proteomics 9, 4048–4062 [DOI] [PubMed] [Google Scholar]
- 38. Karess R. (2005) Rod-Zw10-Zwilch. A key player in the spindle checkpoint. Trends Cell Biol. 15, 386–392 [DOI] [PubMed] [Google Scholar]
- 39. Kops G. J., Kim Y., Weaver B. A., Mao Y., McLeod I., Yates J. R., 3rd, Tagaya M., Cleveland D. W. (2005) ZW10 links mitotic checkpoint signaling to the structural kinetochore. J. Cell Biol. 169, 49–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Salina D., Enarson P., Rattner J. B., Burke B. (2003) Nup358 integrates nuclear envelope breakdown with kinetochore assembly. J. Cell Biol. 162, 991–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stelzl U., Worm U., Lalowski M., Haenig C., Brembeck F. H., Goehler H., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S., Timm J., Mintzlaff S., Abraham C., Bock N., Kietzmann S., Goedde A., Toksöz E., Droege A., Krobitsch S., Korn B., Birchmeier W., Lehrach H., Wanker E. E. (2005) A human protein-protein interaction network. A resource for annotating the proteome. Cell 122, 957–968 [DOI] [PubMed] [Google Scholar]
- 42. Wang X., Babu J. R., Harden J. M., Jablonski S. A., Gazi M. H., Lingle W. L., de Groen P. C., Yen T. J., van Deursen J. M. (2001) The mitotic checkpoint protein hBUB3 and the mRNA export factor hRAE1 interact with GLE2p-binding sequence (GLEBS)-containing proteins. J. Biol. Chem. 276, 26559–26567 [DOI] [PubMed] [Google Scholar]
- 43. Babu J. R., Jeganathan K. B., Baker D. J., Wu X., Kang-Decker N., van Deursen J. M. (2003) Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J. Cell Biol. 160, 341–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sowa M. E., Bennett E. J., Gygi S. P., Harper J. W. (2009) Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hutchins J. R., Toyoda Y., Hegemann B., Poser I., Hériché J. K., Sykora M. M., Augsburg M., Hudecz O., Buschhorn B. A., Bulkescher J., Conrad C., Comartin D., Schleiffer A., Sarov M., Pozniakovsky A., Slabicki M. M., Schloissnig S., Steinmacher I., Leuschner M., Ssykor A., Lawo S., Pelletier L., Stark H., Nasmyth K., Ellenberg J., Durbin R., Buchholz F., Mechtler K., Hyman A. A., Peters J. M. (2010) Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science 328, 593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rahmani Z., Gagou M. E., Lefebvre C., Emre D., Karess R. E. (2009) Separating the spindle, checkpoint, and timer functions of BubR1. J. Cell Biol. 187, 597–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Michel L. S., Liberal V., Chatterjee A., Kirchwegger R., Pasche B., Gerald W., Dobles M., Sorger P. K., Murty V. V., Benezra R. (2001) MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature 409, 355–359 [DOI] [PubMed] [Google Scholar]
- 48. Chung E., Chen R. H. (2002) Spindle checkpoint requires Mad1-bound and Mad1-free Mad2. Mol. Biol. Cell 13, 1501–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Trachana V., van Wely K. H., Guerrero A. A., Fütterer A., Martínez-A C. (2007) Dido disruption leads to centrosome amplification and mitotic checkpoint defects compromising chromosome stability. Proc. Natl. Acad. Sci. U.S.A. 104, 2691–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.