Background: Cyclic nucleotide specific phosphodiesterases (PDEs) are essential enzymes in many parasitic protozoa and represent important new drug targets.
Results: The crystallographic structure and enzymatic properties of Trypanosoma cruzi phosphodiesterase C (TcrPDEC) have been determined.
Conclusion: A parasite-specific pocket next to the TcrPDEC active site might allow designing parasite-specific inhibitors.
Significance: The findings highlight the potential of PDEs as anti-parasite drug targets.
Keywords: Crystal Structure, Cyclic AMP (cAMP), Cyclic GMP (cGMP), Kinetics, Parasite, Phosphodiesterases, Trypanosome
Abstract
Trypanosoma cruzi phosphodiesterase C (TcrPDEC) is a potential new drug target for the treatment of Chagas disease but has not been well studied. This study reports the enzymatic properties of various kinetoplastid PDECs and the crystal structures of the unliganded TcrPDEC1 catalytic domain and its complex with an inhibitor. Mutations of PDEC during the course of evolution led to inactivation of PDEC in Trypanosoma brucei/Trypanosoma evansi/Trypanosoma congolense, whereas the enzyme is active in all other kinetoplastids. The TcrPDEC1 catalytic domain hydrolyzes both cAMP and cGMP with a Km of 23.8 μm and a kcat of 31 s−1 for cAMP and a Km of 99.1 μm and a kcat of 17 s−1 for cGMP, thus confirming its dual specificity. The crystal structures show that the N-terminal fragment wraps around the TcrPDEC catalytic domain and may thus regulate its enzymatic activity via direct interactions with the active site residues. A PDE5 selective inhibitor that has an IC50 of 230 nm for TcrPDEC1 binds to TcrPDEC1 in an orientation opposite to that of sildenafil. This observation, together with the screen of the inhibitory potency of human PDE inhibitors against TcrPDEC, implies that the scaffold of some human PDE inhibitors might be used as the starting model for design of parasite PDE inhibitors. The structural study also identified a unique parasite pocket that neighbors the active site and may thus be valuable for the design of parasite-specific inhibitors.
Introduction
Chagas disease, also known as American trypanosomiasis, is a tropical parasitic disease that currently affects ∼10 million people in Latin America (1, 2). The disease is caused by Trypanosoma cruzi, a kinetoplastid protozoan that is delivered by blood-sucking bedbugs such as Rhodnius prolixus (3). After an acute phase of infection, with a mortality rate of ∼10%, the infection can persist without overt clinical symptoms for 20–30 years. Although a considerable proportion of the cases never progress beyond this initial stage, many patients finally succumb to the slow but inexorable destruction of the musculature of heart, intestines, and esophagus (2).
In the kinetoplastids, including T. cruzi, differentiation and cell cycle regulation are under the control of cyclic nucleotide signaling (4–6), in which phosphodiesterases (PDEs)4 are the sole enzymes that decompose cyclic nucleotides. PDE activity is also involved in the osmoregulation of cell volume after hypoosmotic shock (7, 8). Disruption of cyclic nucleotide signaling has been shown to eliminate Trypanosoma brucei infection from the host (9). In this context, PDE inhibitors might constitute a new class of potent drug candidates for protozoal infections. In T. cruzi, several cyclic nucleotide-specific PDEs have already been characterized (10–14), and a number of TcrPDE inhibitors have been reported (15).
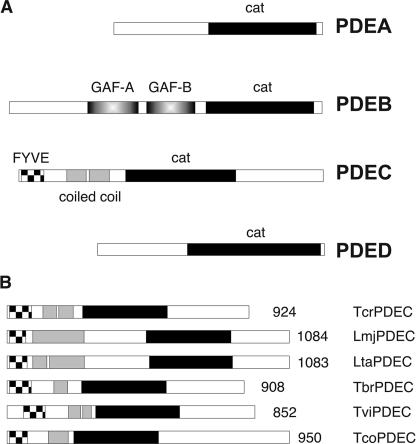
The genomes of kinetoplastids, including T. cruzi, encode four PDE families, termed TcrPDEA, B, C, and D (16) (Fig. 1), in contrast to the human genome, which contains 21 PDE genes categorized into 11 families. TcrPDEA, C, and D each consist of a single member, whereas TcrPDEB contains two highly similar and tandemly arranged genes. TcrPDEA is a cAMP-specific PDE with a high Km and is not inhibited by isobutylmethylxanthine, papaverine, or theophylline (10). The highly similar TcrPDEB1 and TcrPDEB2 are cAMP-specific and contain two GAF domains upstream of their catalytic domains (11, 14, 17). TcrPDEC contains a FYVE-type domain (18) at its N terminus and was shown to hydrolyze both cAMP and cGMP with similar Km values (13). This observation is supported by recent work of Docampo and co-workers (15) but is in contrast to the claim that TcrPDEC is cAMP-specific (11). TcrPDEC activity has been implicated in osmoregulation (8, 12, 15). TcrPDED and its orthologs in other kinetoplastids have so far not been experimentally investigated.
FIGURE 1.
The PDE genes of the kinetoplastids. A, domain structures of the four PDE families that are present in all currently sequenced kinetoplastid genomes (T. brucei, T. cruzi, T. vivax, T. congolense, T. evansi, Crithidia fasciculata, Leishmania tarentolae, L. major, L. braziliensis, L. mexicana, and L. infantum). B, domain structures of PDECs from various kinetoplastid species. Cross-hatched bars, FYVE-type domains; gray bars, coiled-coil regions; black bars, catalytic domains.
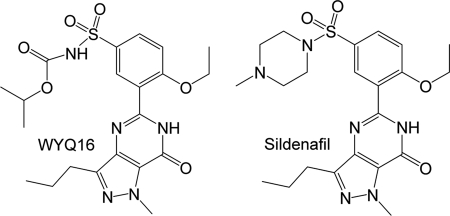
Although Chagas disease and its causative agent T. cruzi were discovered ∼100 years ago (2), there are only a handful of clinically useful drugs at present, and no vaccine is available (19–22). Worse still, all current drugs cause severe side effects, and their use is hampered by drug resistance (23), emphasizing an urgent need for the discovery of new and more effective drugs for the treatment of Chagas disease (24). In this study, we report the enzymatic properties of various kinetoplastid PDECs, and the crystal structures of the unliganded TcrPDEC1 and its complex with the inhibitor wyq16 (5-(2-ethoxy-5-(N-isopropyloxycarbonyl-aminosulfonyl)-1-methyl-3-propyl-1H-pyrazolo[4,3-d]pyrimidin-7(6H)-one) (Fig. 2). These results provide guidelines for the design of TcrPDEC inhibitors for the treatment of Chagas disease.
FIGURE 2.
Chemical structures of sildenafil and wyq16.
EXPERIMENTAL PROCEDURES
Cloning of Kinetoplastid PDEC Genes and Expression in Saccharomyces cerevisiae
The entire PDEC coding sequences of five kinetoplastid species were amplified by PCR (Expand High Fidelity® PCR system; Roche Applied Science) and cloned using a TA cloning kit (TOPO TA® cloning kit from Invitrogen or pGEM®-T easy vector system from Promega). PCR amplification of the respective DNA fragments was performed using the following primer pairs: oPREM-for1 5′-CAGTCGACATATGGTGTGCTACGACGGTTC-3′ and oPREM-rev7b 5′-GAGGATCCTCAAGGACCCACCGACGC-3′ for T. brucei TbrPDEC (genomic DNA of procyclic strain 427), oTconPDE4-for 5′-CAGTCGACATATGTGTAAATACCTTAGCACC-3′ and oTconPDE4-rev 5′-CTGGATCCCTACTTTGCCAACAACGTTGCC-3′ for Trypanosoma congolense TcoPDEC (genomic DNA of strain Savannah ALME/CAM), oTviPDEC-f 5′-GCACATATGTACAGTTCTAAGGAGTCGCAC-3′ and oTviPDEC-r 5′-GCAGATCTTTATCTGTCCAAGAATAATCCTGAG-3′ for Trypanosoma vivax TviPDEC (genomic DNA of strain Y486), oTcPDE4-for 5′-CAGTCGACATATGTCGGAGGACGCTGGGCTTC-3′ and oTcPDE4-r 5′-CGTGGATCCTCAGCACTGCGTCAACAGAGTG-3′ for T. cruzi (genomic DNA of strain Tulahuen), LmjPDEC-for 5′-CAGTCGACATATGTCTCTCATGCCCGGGTACC-3′ and LmjPDEC-r 5′-CTGGATCCTCACATGTACCGCAGTGCAGACG-3′ for Leishmania major (genomic DNA of strain MHRO/IR/75/ER). All of the cloned PCR products were verified by sequencing.
For expression in S. cerevisiae, the entire PDEC open reading frames were subcloned from the TA vectors into two variants of the yeast expression vector pLT1 (13) using the restriction enzymes SalI/BamHI or NdeI/BglII (sites are indicated by bold letters in the primer sequences). One pLT1 variant directs the expression of the kinetoplastid protein alone, whereas the other adds an N-terminal hemagglutinin tag to allow detection of the recombinant protein. Transformation of the constructs into the PDE-deficient S. cerevisiae strain PP5 (MATa leu2-3 leu2-112 ura3-52 his3-532 his4 cam pde1::URA3 pde2::HIS3) was carried out as described previously (25).
Yeast Cell Lysis
Yeast cell lysates were prepared as previously described (26). Briefly, yeast was grown in SC-Leu medium to end log phase, collected by centrifugation, and resuspended in the original volume of prewarmed YPD. The culture was incubated for an additional 3.5 h to maximize protein expression. The cells were then washed twice in H2O, and the pelleted cell was snap-frozen in liquid nitrogen and stored at −70 °C. The pellet was thawed on ice and resuspended in an equal volume of ice-cold extraction buffer (50 mm Hepes, pH 7.5, 100 mm NaCl, 1× Complete® protease inhibitor mixture without EDTA (Roche Applied Science)). Cell lysates were produced by mechanical disruption with glass beads (0.45–0.50 mm) using a FastPrep FP120 high speed reciprocating device (four times for 45 s at setting 4). The lysate was cleared by centrifugation and frozen for later use.
Subcloning of Catalytic Domain TcrPDEC1 in Escherichia coli and Protein Purification
A pair of primers that contain the restriction sites NheI and EcoRI was designed for the subcloning. The DNA sequence encoding the catalytic domain of TcrPDEC1 (amino acids 270–614) was amplified by PCR. The amplified TcrPDEC1 fragment and the expression vector pET28a were digested with the restriction enzymes, purified by agarose gel, and ligated with T4 DNA ligase. The resultant plasmid pET-PDEC1 was transferred into E. coli strain BL21(CodonPlus) (Agilent Technologies) for expression. The E. coli cell carrying pET-PDEC1 was grown in LB medium at 37 °C to an absorption of A600 = 0.7, and then 0.1 mm isopropyl β-d-thiogalactopyranoside was added to further grow at 15 °C for 24 h. The harvested cells were suspended in a lysis buffer (3 ml/gram) consisting of 20 mm Tris-HCl, pH 7.5, 300 mm NaCl, and 15 mm imidazole and disrupted by French Press with 1500 p.s.i. After centrifugation at 15,000 rpm for 20 min, the supernatant was loaded into a nickel-nitrilotriacetic acid column (Qiagen). The nickel-nitrilotriacetic acid column was washed with ∼300 ml of lysis buffer and then ∼100 ml of buffer of 20 mm Tris-HCl, pH 8,0, 50 mm NaCl, 15 mm imidazole. The TcrPDEC1 protein was eluted by using 20 mm Tris-HCl, pH 7.5, 50 mm NaCl, and 250 mm imidazole. The N-terminal His6-tagged TcrPDEC1 was cleaved by thrombin, and the protein was further purified by Q-Sepharose with three buffers of 20 mm Tris-HCl, pH 7.5, 1 mm 2-mercaptoethanol, 1 mm EDTA, and NaCl at 0.1, 0.2, and 0.3 m. Finally, a Sephacryl S300 column (GE Healthcare) was used to remove aggregated protein and to switch to a buffer consisting of 20 mm Tris-HCl, pH 7.5, 1 mm 2-mercaptoethanol, 1 mm EDTA, 50 mm NaCl. A typical purification yielded >10 mg of TcrPDEC1 with a purity >95% from a 2-liter cell culture. Protein concentrations were calculated from extinction coefficients by the program ProtParam, and an A280 of 0.701 m−1 cm−1 was equal to 1 mg/ml of TcrPDEC1 (residues 270–614).
Selenomethionyl-substituted TcrPDEC1 was prepared using the protocol described by Doublie (27). The E. coli cell was cultured in the M9 minimal medium that was supplemented with the following amino acids: 100 mg/ml Lys, 100 mg/ml Thr, 100 mg/ml Phe, 50 mg/ml Leu, 50 mg/ml Ile, 50 mg/ml Val, and 50 mg/ml selenomethionine. The protocol for purification of selenomethionyl-substituted TcrPDEC1 was the same as that used for purification of the wild-type TcrPDEC1.
Enzymatic Properties
Enzymatic activities were assayed using [3H]cAMP and [3H]cGMP as substrates, as previously reported (28). The catalytic domain of TcrPDEC1 was incubated at 24 °C for 15 min with a reaction mixture of 20 mm Tris·HCl, pH 7.5, 10 mm MgCl2, 1 mm DTT, [3H]AMP, or [3H]cGMP (20,000–40,000 cpm/assay, ∼20 nm in a 100-μl assay), and appropriate concentrations of nonradiolabeled cAMP or cGMP. The reaction was terminated by the addition of 0.2 m ZnSO4. The reaction product [3H]AMP or [3H]GMP was precipitated out by the addition of 0.25 m Ba(OH)2, whereas unreacted [3H]AMP or [3H]cGMP remained in the supernatant. After centrifugation, the radioactivity in the supernatant was measured in a liquid scintillation counter. The turnover was controlled at hydrolysis of 15–30% substrate under a suitable enzyme concentration. Nine concentrations of cAMP or cGMP in a range of 0.04–800 μm were used to obtain the kinetic parameters.
Enzymatic properties were analyzed by steady state kinetics, as shown in the form of Michealis-Menten's equation (29). The nonlinear regression (GraphPad Prism 5.0) was applied to obtain the values of Km, Vmax, and kcat. For measurement of IC50, eight concentrations of inhibitors were used at a substrate concentration of <1/10 Km and a suitable enzyme concentration. All of the measurements were repeated three times.
For screening potential inhibitors, a set of human PDE inhibitors was dissolved in Me2SO as 10 mm stocks and used at a concentration of 100 μm in the PDE assay. The assay was conducted with lysates of yeast expressing full-length TcrPDEC1 as described (26). 1 μm cAMP was used as substrate, and no more than 20% substrate was hydrolyzed in all reactions. The Me2SO concentration in the final assay solutions never exceeded 1%, and appropriate control reactions with Me2SO alone were always included.
Crystallization and Structure Determination of TcrPDEC1
The catalytic domain of the unliganded TcrPDEC1 (270–614) and its complex with wyq16 was crystallized by vapor diffusion against a well buffer of 20% PEG 3350, 0.4 m sodium formate, 0.2 m guanidine, 0.1 m MES, pH 6.5, at 4 °C. The hanging drops contain 2 μl of protein (10 mg/ml) and 2 μl of well buffer. The inhibitor complex was prepared by incubation of TcrPDEC1 with 2 mm wyq16 (0.1 m stock in Me2SO) for 1 h on ice before crystallization. Diffraction data were collected on Beamline X29 at Brookhaven National Laboratory (Table 1) and processed by the program HKL (30). The crystal of the unliganded TcrPDEC1 has the space group P41212 with cell dimensions of a = b = 130.3 Å and c = 388.9 Å. The TcrPDEC1-wyq16 complex was crystallized in the same space group but slightly different cell dimensions of a = b = 131.1 Å and c = 394.9 Å.
TABLE 1.
Statistics of diffraction data and structure refinement
| Native TcrPDEC1 | TcrPDEC1-wyq16 | Se-TcrPDEC1 | |
|---|---|---|---|
| Data collection | |||
| Space group | P41212 | P41212 | P41212 |
| Unit cell (a, c) (Å) | 130.3, 388.9 | 131.1, 394.7 | 131.2, 395.1 |
| Resolution range (Å) | 30.0–2.0 | 30.0–2.33 | 30.0–2.33 |
| Unique reflections | 219, 103 | 146, 141 | 278, 015 |
| Fold of redundancy | 8.9 | 27.2 | 14.3 |
| Completeness (%) | 94.4 (100.0)a | 98.7 (100.0)a | 98.6 (100.0)a |
| Average I/σ | 8.4 (4.2)a | 8.3 (5.0)a | 5.8 (4.8)a |
| Rmerge | 0.101 (0.61)a | 0.125 (0.60)a | 0.156 (0.80)a |
| Structure refinement | |||
| Rfactor | 0.216 | 0.227 | |
| Rfree | 0.246 (10%)b | 0.267 (10%)b | |
| Resolution range (Å) | 30.0–2.0 | 30.0–2.33 | |
| Reflections | 205, 388 | 140, 293 | |
| Root mean square deviation | |||
| For bond length | 0.0055 | 0.0089 | |
| For angle | 1.11° | 1.55° | |
| Average B factor (Å2) | |||
| Protein | 30.1 (20520)c | 32.5 (20633)c | |
| wyq16 | 37.7 (264)c | ||
| Water | 27.0 (975)c | 28.1 (874)c | |
| Zn2+ | 23.2 (8)c | 27.4 (8)c | |
| Mg2+ | 22.1 (8)c | 23.6 (8)c | |
| No. of molecules in the asymmetric unit | 8 | 8 | |
| No. of disallowed residues in Ramachandran plot | 0 | 0 | |
a The numbers in parentheses are for the highest resolution shells, 2.07–2.0 for the native and 2.41–2.33 Å for the wyq16 complex.
b 10% of reflections were omitted in the refinement for calculation of Rfree.
c The number of atoms in the crystallographic asymmetric unit.
The structure of TcrPDEC1 was solved by the single-wavelength anomalous diffraction method (31), using a selenomethionine derivative of the TcrPDEC1 catalytic domain. The initial positions of selenium atoms were located by program SHELX-97 (32) and refined to yield a figure of merit of 0.43. The map was subsequently modified by program PHENIX (33) and clearly revealed the protein trace (supplemental Fig. S1). The structure of TcrPDEC1 in complex with inhibitor wyq16 was solved by the molecular replacement program AMoRe (34), using the native TcrPDEC1 structure as the initial model. The phases from the molecular replacement were improved by the density modification package of CCP4. The structures were rebuilt by the program O (35) and refined by the program CNS (36).
RESULTS AND DISCUSSION
PDEC Is Inactive in Some Kinetoplastid Species
Sequence analysis of the kinetoplastid genomes revealed the presence of a PDEC gene in each of these genomes: T. cruzi (Tc00.1047053506697.20, chromosome 36-S), T. brucei (Tb927.3.3070, chromosome 3), and Trypanosoma evansi.5 The sequences of TbrPDEC and TevPDEC differ by only three amino acids, one of which is within the catalytic domain (Ala-598 of T. brucei versus Val-599 in T. evansi.). T. congolense TcoPDEC gene was sequenced from two different Savannah strains, ALME/CAM and CARBA/CAM. These two sequences were identical but differed from the TcIL3000.3.1980 (chromosome 3) reference sequences deposited in GeneDB (42/2851 nucleotides, 13/950 amino acids): T. vivax (chromosome 3),6 Leishmania infantum (LinJ.29.2790, chromosome 29), Leishmania mexicana (LmxM08_29.2680, chromosome 29), Leishmania braziliensis (LbrM.29.2740, chromosome 29), and L. major (LmjF.29.2680, chromosome 29). Unexpectedly, the PDEC sequence alignment (supplemental Fig. S2) showed that two histidines conserved in all class 1 PDEs (37) are replaced with Arg-369 and Leu-406 in T. brucei and T. evansi and with Arg-421 and Leu-458 in T. congolense. Furthermore, a conserved hydrophobic residue in class 1 PDEs (methionine, leucine, or isoleucine) is replaced by either threonine (Thr-479 in T. brucei and T. evansi) or tyrosine (Tyr0531 in T. congolense). Because these residues are critical for binding of the metal ion and substrate, the mutations would presumably inactivate the PDEs. To verify this prediction, the PDECs of T. brucei and T. congolense (predicted to be inactive) and of T. vivax, T. cruzi, and L. major (predicted to be active) were expressed in a PDE-deficient strain of S. cerevisiae. Complementation of PDE-deficient S. cerevisiae restores its heat shock resistance and represents a very sensitive assay for cAMP-PDE activity (25). As predicted by sequence analysis, the PDECs of T. cruzi, L. major, and T. vivax were catalytically active and able to complement the PDE deficiency (Fig. 3A). In contrast, the PDEC of neither T. brucei nor T. congolense was active in this assay (Fig. 3A), although the expression of their recombinant proteins was ascertained by immunoblotting (Fig. 3, B and C). Phylogenetic analysis demonstrated that the mutational inactivation of PDEC occurred rather late in the evolution of the trypanosomes, after the separation of the T. brucei/T. congolense/T. evansi clade from the remainder of the trypanosomatids (Fig. 3D) (38).
FIGURE 3.
Determination of PDEC activity by complementation of a PDE-deficient S. cerevisiae strain. A, heat shock assay on the strains expressing PDEC from various trypanosomatids. Left panel, control incubation without heat shock; right panel, 15-min heat shock (55 °C) before incubating the plate at 30 °C. −HA or +HA, expression constructs without or with an N-terminal hemagglutinin tag. Both constructs are similarly effective for complementation. Negative control is the yeast strain transfected with empty vector. B, PDEC expression in the recombinant yeast strains. Recombinant PDECs were detected by immunoblotting with anti-hemagglutinin antibody. C, Ponceau staining. D, kinetoplastid phylogeny derived from maximal likelihood of 18 S ribosomal RNA (adapted from Ref. 26), confirming the split of T. vivax from T. brucei. Gray boxes, species with inactive PDEC.
The fact that the open reading frames of TbrPDEC, TviPDEC, and TevPDEC encode full-length but catalytically inactive proteins implies that these proteins play a role unrelated to their PDE activity. To determine whether inactive PDECs are expressed, transcription of PDEC was studied in T. brucei. Deep sequencing of mRNAs of various stages of the T. brucei life cycle (39) revealed that stable transcripts of the TbrPDEC gene (Tb927.3.3070) are present in all life cycle stages and that they are processed at one major and a few minor splice sites. The overall abundance of TbrPDEC transcripts in T. brucei (∼50/106 total transcripts) is not significantly different between different life cycle stages (bloodstream versus procyclic forms). It is also similar to that of the enzymatically active TcrPDEC of T. cruzi (∼30/106 total transcripts).7,8 Northern blot analysis indicates a transient 2-fold increase in mRNA levels during transformation from bloodstream to procyclic forms, in agreement with a recent microarray analysis that demonstrated a similar up-regulation of Tb927.3.3070 during differentiation from short stumpy to procyclic forms (40). The expression of the TbrPDEC protein has also been established in a proteomics approach with procyclic T. brucei (41) (the older designation Tb03.27C5.640 was used in this work).
Kinetics of TcrPDEC1 Catalytic Domain
Two alleles of TcrPDEC from T. cruzi have been sequenced and were shown to vary in 35 of 924 amino acids. The TcrPDEC1 catalytic domain (residues 270–614) hydrolyzes cAMP with a Km of 23.8 μm and a kcat of 31 s−1 and cGMP with a Km of 99.1 μm and a kcat of 17 s−1. The catalytic efficiency kcat/Km is 1.3 and 0.18 s−1 μm−1, respectively, for cAMP and cGMP and indicates the dual specificity of TcrPDEC1 with ∼7-fold preference of cAMP over cGMP (Table 2). These enzymatic parameters are consistent with the Km values of 31.6 and 78.2 μm for cAMP and cGMP, which were reported earlier for full-length TcrPDEC1 (13). It is interesting to note that the kcat of the TcrPDEC1 catalytic domain is 7.5–90 times the kcat values of other reported PDEs, although the catalytic efficacy of TcrPDEC1 is still comparable with other PDEs because of its weak apparent association constant (Table 2).
TABLE 2.
Kinetic parameters of PDEs
The kinetics data of LmjPDEB1 and human PDEs are cited from Refs. 28, 43, 45, 52, and 53. ND, not determined.
| cAMP |
cGMP |
|||||
|---|---|---|---|---|---|---|
| Km | kcat | kcat/Km | Km | kcat | kcat/Km | |
| μm | s−1 | s−1μm−1 | μm | s−1 | s−1μm−1 | |
| TcrPDEC (270–614) | 21.3 ± 2.1 | 30.7 ± 0.9 | 1.4 ± 0.1 | 87.0 ± 6.2 | 16.7 ± 0.4 | 0.19 ± 0.02 |
| LmjPDEB1 (582–940) | 20.6 ± 3.4 | 2.7 ± 0.3 | 0.13 ± 0.01 | >1000 | ND | ND |
| PDE4D2 | 1.5 ± 0.2 | 3.9 ± 0.3 | 2.7 ± 0.4 | 990 ± 100 | 5.2 ± 0.8 | 0.053 ± 0.008 |
| PDE5A1 (535–860) | ND | ND | ND | 5.1 ± 1.3 | 1.3 ± 0.3 | 0.27 ± 0.08 |
| PDE7A1 (130–482) | 0.20 ± 0.03 | 1.6 ± 0.2 | 7.9 ± 0.9 | 3900 ± 700 | 6.8 ± 1.3 | 0.0018 ± 0.0001 |
| PDE8A1 (480–820) | 1.8 ± 0.1 | 4.0 ± 0.1 | 2.2 ± 0.1 | 1600 ± 100 | 1.6 ± 0.2 | 0.001 ± 0.0001 |
| PDE10A2 (449–789) | 0.056 ± 0.005 | 0.33 ± 0.02 | 5.9 ± 0.6 | 4.4 ± 0.3 | 1.2 ± 0.1 | 0.27 ± 0.01 |
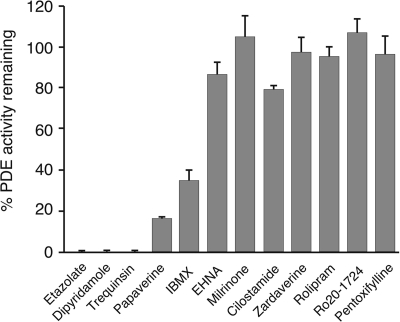
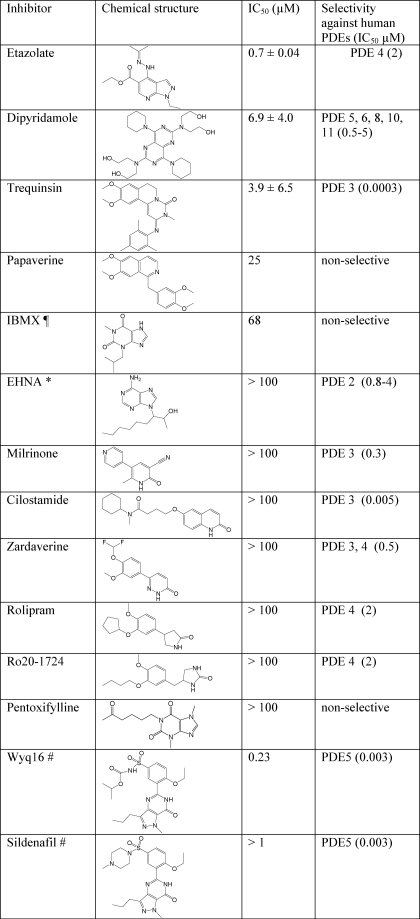
The potency of various inhibitors of human PDEs was assayed against full-length TcrPDEC1 (Fig. 4 and Table 3). It was found that some inhibitors such as etazolate and trequinsin are very potent (Table 3 for IC50 values), and thus their scaffolds might be taken as starting models for the design of TcrPDEC selective inhibitors.
FIGURE 4.
Potency of human PDE inhibitors against TcrPDEC1. Compounds were tested at 100 μm against full-length recombinant TcrPDEC1.
TABLE 3.
Potency of human PDE inhibitors against TcrPDEC
¶ IBMX, 3-isobutyl-1-methylxanthine.
* EHNA, erythro-9-(2-hydroxy-3-nonyl)-adenosine.
# The IC50 values of these compounds were measured by using the TcrPDEC1 catalytic domain.
Structure of TcrPDEC1 Catalytic Domain
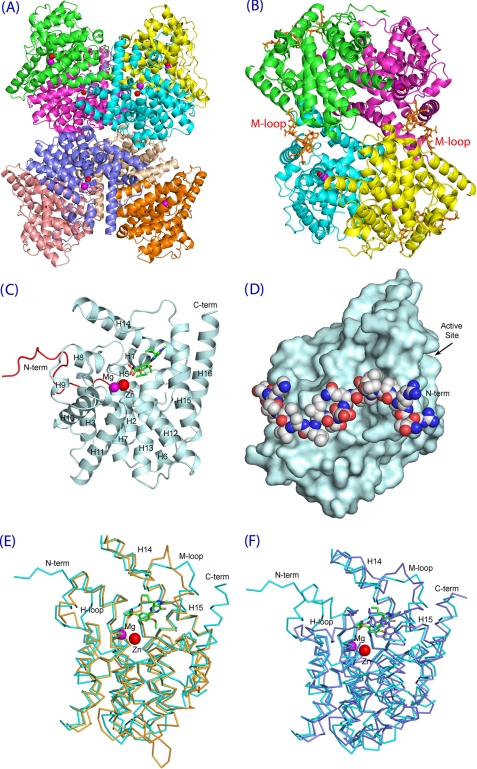
The structures of the unliganded TcrPDEC1 catalytic domain (traceable residues 277–609) and its complex with inhibitor wyq16 were determined at medium resolution (Table 1). The crystallographic asymmetric unit contains eight molecules that are associated into an octamer or two tetramers (Fig. 5, A and B). The oligomerization of the TcrPDEC1 catalytic domain is different from those of human PDEs and LmjPDEB1 (42, 43). It is not clear whether the tight tetrameric or octameric association of the TcrPDEC1 catalytic domain is biologically relevant.
FIGURE 5.
The structure of the TcrPDEC1 catalytic domain. A, ribbon diagram of octamer of the TcrPDEC1 catalytic domain. B, ribbon of tetrameric TcrPDEC1. C, ribbon of TcrPDEC1 monomer. The inhibitor wyq16 is presented as the ball and stick model. The red coil represents the long N-terminal fragment that stretches from the back to the front of the molecule and approaches the active site. D, surface presentation of the TcrPDEC1 catalytic domain, in a view of ∼100° rotation around the vertical axis of C. The balls represent the long N-terminal fragment that stretches around the domain. E, superposition between the catalytic domains of TcrPDEC1 (cyan) and human PDE4D2 (yellow). F, superposition between TcrPDEC1 (cyan) and human PDE5A1 (light purple). The figure shows that the M-loops have significant conformational differences.
A monomer of the TcrPDEC1 catalytic domain comprises 16 α-helices with two divalent metal ions bound at the bottom of the active site (Fig. 5, C and D). The α-helices are arranged in a similar topology as those of the catalytic domains of human PDEs and LmjPDEB1 (42, 43). The nature of the divalent metal ions was not identified, but zinc and magnesium were used in the structure refinement, as it has been done in the structures of human PDEs (42). The metal ion assignment appears to be reasonable and is supported by the comparable B-factors of the metal ions with the protein residues (Table 1) and proper electron density in the unbiased maps of (Fo − Fc) and (2Fo − Fc). The zinc ion chelates with His-372, His-409, Asp-410, and Asp-521, plus two water molecules in an octahedral configuration. The magnesium ion coordinates with Asp-410 and five water molecules to form an octahedron. An unexpected observation of this study is that the N-terminal fragment of TcrPDEC1 stretches from the back to the front of the molecule and approaches the substrate-binding pocket of the enzyme (Fig. 5, C and D). This observation implies that the N terminus might play a role in regulation of the enzymatic activity of TcrPDEC1 via a direct interaction with the active site. This argument is consistent with the report that a fragment of the upstream conserved region of human PDE4 interacts with the inhibitors at the active site (44). However, the corresponding loop of PDE4, which links the upstream conserved region fragment to the back of the PDE4 domain, is disordered, in contrast to the observable conformation of our N-terminal stretch.
Structural superposition of the TcrPDEC1-wyq16 complex over human PDE4D, human PDE5A, and LmjPDEB1 yielded RMS deviations of 1.45, 1.38, and 1.59 Å for the Cα atoms of 269, 287, and 247 comparable residues, respectively (Fig. 5, E and F), indicating their overall similarity. However, there are some subtle but significant differences between them. TcrPDEC1 lacks the short helix H4 but has an extra helix before H7. The long helix H14 is split into two helices in TcrPDEC1 because of a missing amino acid before Val-537. The biological meaning of these differences is not clear but might simply reflect the consequence of the amino acid sequence variation. The most significant change is associated with the M-loop (residues 544–564) at the active sites of the PDEs, which showed differences of several Angstroms between their Cα atoms (Fig. 5, B, E, and F). These conformational changes do not appear to result from inhibitor binding, as shown by the comparable conformations and positions of the M-loops between the unliganded and wyq16-bound structures. However, it is not clear whether the M-loop changes represent the native conformation differences between TcrPDEC and other PDEs or the consequence of the crystal packing because some residues of the M-loop such as Pro-558 and Glu-561 are involved in formation of the tetramer in the crystal (Fig. 5B). On the other hand, the H-loop at the active site of TcrPDEC favorably compares with that of human PDE4 but not PDE5. Because most human PDE families have an H-loop conformation similar to PDE4 and PDE5 uniquely shows multiple conformations (42, 45, 46), the H-loop might not be an attractive target for the design of TcrPDEC inhibitors.
Inhibitor Binding
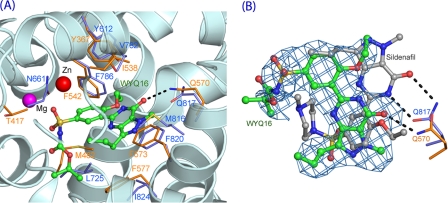
The PDE5 inhibitor wyq16 (Fig. 2) shares the ethoxyphenyl and pyrazolopyrimidinone groups with sildenafil but differs in an isopropyloxycarbonylamino group from N-methylpiperazinyl. wyq16 inhibits the catalytic activities of PDE5A and TcrPDEC1 with IC50 values of 3.2 and 230 nm, respectively. The electron density maps clearly define the conformation and position of the ethoxyphenyl and pyrazolopyrimidinone groups of wyq16 (supplemental Fig. S3). However, the isopropyloxycarbonylamino tail seems more flexible, as shown by the weaker density and higher B-factors. wyq16 binds to the TcrPDEC1 active site with two conserved characteristics: the NE2 atom of the invariant Gln-570 forms a hydrogen bond with O1 of wyq16, and Phe-573 stacks against the pyrazolopyrimidinone ring of wyq16 (Fig. 6). In addition, wyq16 forms van der Waals contacts with residues Tyr-367, Thr-417, Met-482, Leu-496, Ile-538, Phe-542, Phe-560, Ser-569, and Phe-577. An unexpected finding is that the common pyrazolopyrimidinone rings of wyq16 and sildenafil orient oppositely. As a result, the hydrogen bonding patterns are completely different, although the pyrazolopyrimidinone rings of both molecules stack against the phenylalanine (Phe-573 in TcrPDEC1). The side chain of the invariant glutamine forms two hydrogen bonds with the pyrazolopyrimidinone ring of sildenafil in PDE5A (45), but only one with wyq16 (Fig. 6B). This difference might be due to the different conformations of the invariant glutamine, in which the glutamine side chain of TcrPDEC1 has an opposite orientation to that of PDE5 (Fig. 6) but the same as that of PDE4.
FIGURE 6.
Binding of inhibitor wyq16. A, wyq16 binding to the active site of TcrPDEC1. The dotted lines represent hydrogen bonds. The TcrPDEC1 and PDE5A residues are presented as yellow and light blue sticks, respectively. B, superposition of wyq16 (green sticks) over sildenafil (gray). The invariant glutamine (Gln-570 of TcrPDEC1 and Gln-817 of PDE5A) have different orientations of their side chains. The light blue nets represent the (Fo − Fc) electron density that was calculated from the TcrPDEC1 structure with omission of wyq16 and contoured at 2.0.
Potential Parasite Pocket
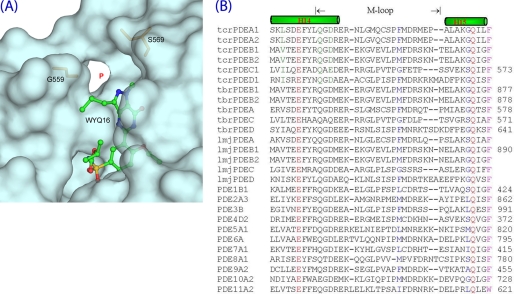
Our earlier work revealed that LmjPDEB1 has a small pocket neighboring the active site (originally designated as the L-pocket) (43). The TcrPDEC1 structures further showed that the pocket is unique and universal for the kinetoplastid PDEs, and thus the term “P-pocket” appears to be more appropriate. The P-pocket is made up of residues from the M-loop and helix H14, including Leu-539, Ala-543, Gly-559, Ser-564, Val-566, and Ser-569 of TcrPDEC1 (Fig. 7A). Two key residues, Gly-559 and Ser-569, serve as the gatekeepers guarding the access to the P-pocket. In most other kinetoplastid PDE families, the residues corresponding to Gly-559 and Ser-569 are methionine/phenylalanine and glycine (Fig. 7B). Therefore, the gate to the P-pocket of all the kinetoplastid PDEs is sterically open and accessible. However, in human PDEs, this pocket is either blocked by two large gating residues or disappears because of variations in sequence, as shown by the sequence alignment (Fig. 7B). Thus, the P-pocket might be an invaluable structural element for developing parasite-specific PDE inhibitors.
FIGURE 7.
The parasite pocket (P-pocket). A, P-pocket of TcrPDEC1 with bound wyq16. Residues Glt-559 and Ser-569 of TcrPDEC1 are gatekeepers. B, sequence alignment of the corresponding pocket regions in trypanosome, leishmania, and human PDEs. Two gatekeeper residues (blue) isolate the P-pocket from the active sites in human PDE families.
Hint on Use of Some Human PDE Inhibitors as Starting Scaffolds for Design of Parasite PDE Inhibitors
The kinetic and structural studies imply that certain inhibitors of human PDEs could be used as the starting framework for the design of parasite-selective inhibitors. This argument is supported by the inhibitory effects of some human PDE inhibitors on TcrPDEC (Fig. 4 and Table 3). For example, etazolate inhibits human PDE4 and TcrPDEC with the IC50 values of 2 and 0.7 μm, respectively, for the full-length enzymes (13) and of 1.1 and 0.3 μm for the PDE4D2 and TcrPDEC catalytic domains. Moreover, the pyrazolopyrimidinone ring of wyq16 binds to TcrPDEC in an opposite orientation to the same ring of sildenafil in PDE5A (45). Although the IC50 of wyq16 is only 230 nm, the pyrazolopyrimidinone ring might be used as a potential starting scaffold for TcrPDEC inhibitors because of the opposite orientations of both the nine-membered ring and the side chain of the invariant glutamine. Finally, the structure shows that the pyzarole of wyq16 points toward the P-pocket. A suitable substitution on the pyzarole ring would extend the inhibitors to the P-pocket and may thus improve their affinity and selectivity.
Implication of Inactive Parasite PDECs
The finding that mutations during the course of evolution inactivated the PDECs of T. brucei, T. evansi, and T. congolense, has come as a surprise. Interestingly, such a case has also been reported for other enzymes of eukaryotes, including protein kinases (47), protein phosphatases (48), and proteases (49). For example, the EGF receptor kinase HER3 is mutationally locked in an inactive conformation, despite the fact that it can still bind ATP and coordinate Mg2+. However, HER3, rather than serving as a kinase, plays an important role in controlling the activity of other EGF receptors upon ligand binding (50). In another example, inactive homologs of the rhomboid membrane proteases play an important role in controlling the activity of the active enzymes (49).
A systematic survey of metazoan enzymes has identified many inactive forms of other enzymes, but PDE homologs (51). Thus, the kinetoplastid PDECs represent the first case of a PDE family, in which active and inactive homologs coexist and are stably expressed. The inactive TbrPDEC gene is regulated and expressed in T. brucei at similar levels as the active TcrPDEC in T. cruzi, suggesting that the inactive PDECs may still fulfill a cellular function unrelated to cyclic nucleotide hydrolytic activity. Nevertheless, the function of the active enzyme in species of L. major and T. cruzi remains to be established in detail. Of equal interest will be the elucidation of whether maintenance of the inactive PDECs in T. brucei, T. congolense, and T. evansi is due to a selective pressure exerted by their life style.
Supplementary Material
Acknowledgments
We thank Beamline X29 at National Synchrotron Light Source for collection of the diffraction data. We thank Rebecca R. Stanway for proofreading of the manuscript, Torsten Ochsenreiter and Kapila Gunasekera for deep sequencing, Frédéric Bringaud for phylogenetic information, Achim Schnaufer for unpublished T. evansi sequences, Reto Brun, Christiane Hertz-Fowler, and Wendy Gibson for various kinetoplastid cultures and genomic DNAs, and Xuan Lan Vu for outstanding technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant GM59791 (to H. K.). This work was also supported by Grant 3100A-109245 from the Swiss National Science Foundation and TI Pharma Project T4-302 (to T. S.), the Offices of Biological and Environmental Research and of Basic Energy Sciences of the United States Department of Energy, and the National Center for Research Resources of National Institutes of Health (to H. R.).

This article contains supplemental Figs. S1–S3.
The atomic coordinates and structure factors (codes 3V93 and 3V94) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
A. Schnaufer, personal communication.
S. Kunz, unpublished data.
K. Gunasekera, personal communication.
T. Ochsenreiter, personal communication.
- PDE
- phosphodiesterase
- TcrPDEC
- T. cruzi phosphodiesterase C
- wyq16
- 5-(2-ethoxy-5-(N-isopropyloxycarbonyl-aminosulfonyl)-1-methyl-3-propyl-1H-pyrazolo[4,3-d]pyrimidin-7(6H)-one.
REFERENCES
- 1. Rassi A., Jr., Rassi A., Marin-Neto J. A. (2010) Chagas disease. Lancet 375, 1388–1402 [DOI] [PubMed] [Google Scholar]
- 2. Lescure F. X., Le Loup G., Freilij H., Develoux M., Paris L., Brutus L., Pialoux G. (2010) Chagas disease. Changes in knowledge and management. Lancet Infect. Dis. 10, 556–570 [DOI] [PubMed] [Google Scholar]
- 3. Barrett M. P., Burchmore R. J., Stich A., Lazzari J. O., Frasch A. C., Cazzulo J. J., Krishna S. (2003) The trypanosomiases. Lancet 362, 1469–1480 [DOI] [PubMed] [Google Scholar]
- 4. Gould M. K., de Koning H. P. (2011) Cyclic-nucleotide signalling in protozoa. FEMS Microbiol. Rev. 35, 515–541 [DOI] [PubMed] [Google Scholar]
- 5. Laxman S., Beavo J. A. (2007) Cyclic nucleotide signaling mechanisms in trypanosomes. Possible targets for therapeutic agents. Mol. Interv. 7, 203–215 [DOI] [PubMed] [Google Scholar]
- 6. Seebeck T., Schaub R., Johner A. (2004) cAMP signalling in the kinetoplastid protozoa. Curr. Mol. Med. 4, 585–599 [DOI] [PubMed] [Google Scholar]
- 7. Rohloff P., Montalvetti A., Docampo R. (2004) Acidocalcisomes and the contractile vacuole complex are involved in osmoregulation in Trypanosoma cruzi. J. Biol. Chem. 279, 52270–52281 [DOI] [PubMed] [Google Scholar]
- 8. Schoijet A. C., Miranda K., Medeiros L. C., de Souza W., Flawiá M. M., Torres H. N., Pignataro O. P., Docampo R., Alonso G. D. (2011) Defining the role of a FYVE domain in the localization and activity of a cAMP phosphodiesterase implicated in osmoregulation in Trypanosoma cruzi. Mol. Microbiol. 79, 50–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Oberholzer M., Marti G., Baresic M., Kunz S., Hemphill A., Seebeck T. (2007) The Trypanosoma brucei cAMP phosphodiesterases TbrPDEB1 and TbrPDEB2. Flagellar enzymes that are essential for parasite virulence. FASEB J. 21, 720–731 [DOI] [PubMed] [Google Scholar]
- 10. Alonso G. D., Schoijet A. C., Torres H. N., Flawiá M. M. (2007) TcrPDEA1, a cAMP-specific phosphodiesterase with atypical pharmacological properties from Trypanosoma cruzi. Mol. Biochem. Parasitol. 152, 72–79 [DOI] [PubMed] [Google Scholar]
- 11. Díaz-Benjumea R., Laxman S., Hinds T. R., Beavo J. A., Rascón A. (2006) Characterization of a novel cAMP-binding, cAMP-specific cyclic nucleotide phosphodiesterase (TcrPDEB1) from Trypanosoma cruzi. Biochem. J. 399, 305–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Alonso G. D., Schoijet A. C., Torres H. N., Flawiá M. M. (2006) TcPDE4, a novel membrane-associated cAMP-specific phosphodiesterase from Trypanosoma cruzi. Mol. Biochem. Parasitol. 145, 40–49 [DOI] [PubMed] [Google Scholar]
- 13. Kunz S., Oberholzer M., Seebeck T. (2005) A FYVE-containing unusual cyclic nucleotide phosphodiesterase from Trypanosoma cruzi. FEBS J. 272, 6412–6422 [DOI] [PubMed] [Google Scholar]
- 14. D'Angelo M. A., Sanguineti S., Reece J. M., Birnbaumer L., Torres H. N., Flawiá M. M. (2004) Identification, characterization and subcellular localization of TcPDE1, a novel cAMP-specific phosphodiesterase from Trypanosoma cruzi. Biochem. J. 378, 63–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. King-Keller S., Li M., Smith A., Zheng S., Kaur G., Yang X., Wang B., Docampo R. (2010) Chemical validation of phosphodiesterase C as a chemotherapeutic target in Trypanosoma cruzi, the etiological agent of Chagas' disease. Antimicrob. Agents Chemother. 54, 3738–3745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kunz S., Beavo J. A., D'Angelo M. A., Flawia M. M., Francis S. H., Johner A., Laxman S., Oberholzer M., Rascon A., Shakur Y., Wentzinger L., Zoraghi R., Seebeck T. (2006) Cyclic nucleotide specific phosphodiesterases of the kinetoplastida. A unified nomenclature. Mol. Biochem. Parasitol. 145, 133–135 [DOI] [PubMed] [Google Scholar]
- 17. Heikaus C. C., Pandit J., Klevit R. E. (2009) Cyclic nucleotide binding GAF domains from phosphodiesterases. Structural and mechanistic insights. Structure 17, 1551–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lemmon M. A. (2003) Phosphoinositide recognition domains. Traffic 4, 201–213 [DOI] [PubMed] [Google Scholar]
- 19. Seebeck T., Sterk G. J., Ke H. (2011) Phosphodiesterase inhibitors as a new generation of antiprotozoan drugs. Exploiting the benefit of enzymes that are highly conserved between host and parasite. Future Med. Chem. 3, 1289–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Salas C. O., Faúndez M., Morello A., Maya J. D., Tapia R. A. (2011) Natural and synthetic naphthoquinones active against Trypanosoma cruzi. An initial step towards new drugs for Chagas disease. Curr. Med. Chem. 18, 144–161 [DOI] [PubMed] [Google Scholar]
- 21. Castillo E., Dea-Ayuela M. A., Bolás-Fernández F., Rangel M., González-Rosende M. E. (2010) The kinetoplastid chemotherapy revisited. Current drugs, recent advances and future perspectives. Curr. Med. Chem. 17, 4027–4051 [DOI] [PubMed] [Google Scholar]
- 22. Buckner F. S., Navabi N. (2010) Advances in Chagas disease drug development. 2009–2010. Curr. Opin. Infect. Dis. 23, 609–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wilkinson S. R., Kelly J. M. (2009) Trypanocidal drugs. Mechanisms, resistance and new targets. Expert Rev. Mol. Med. 11, e31. [DOI] [PubMed] [Google Scholar]
- 24. de Koning H. P., Gould M. K., Sterk G. J., Tenor H., Kunz S., Luginbuehl E., Seebeck T. (2012) J. Infect. Dis., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kunz S., Kloeckner T., Essen L. O., Seebeck T., Boshart M. (2004) TbPDE1, a novel class I phosphodiesterase of Trypanosoma brucei. Eur. J. Biochem. 271, 637–647 [DOI] [PubMed] [Google Scholar]
- 26. Johner A., Kunz S., Linder M., Shakur Y., Seebeck T. (2006) Cyclic nucleotide specific phosphodiesterases of Leishmania major. BMC Microbiol. 6, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Doublié S. (1997) Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 276, 523–530 [PubMed] [Google Scholar]
- 28. Wang H., Liu Y., Chen Y., Robinson H., Ke H. (2005) Multiple elements jointly determine inhibitor selectivity of cyclic nucleotide phosphodiesterases 4 and 7. J. Biol. Chem. 280, 30949–30955 [DOI] [PubMed] [Google Scholar]
- 29. Fersht A. (1999) in Structure and Mechanism in Protein Science, pp. 103–132, Freeman and Company, New York [Google Scholar]
- 30. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 31. Hendrickson W. A., Ogata C. M. (1997) Phase determination from multiwavelength anomalous diffraction measurements. Methods Enzymol. 276, 494–523 [DOI] [PubMed] [Google Scholar]
- 32. Sheldrick G. M. (2010) Experimental phasing with SHELXC/D/E. Combining chain tracing with density modification. Acta Crystallogr. D Biol. Crystallogr. 66, 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX. A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Navaza J., Saludjian P. (1997) AMoRe: an automated molecular replacement program package. Methods Enzymol. 276, 581–594 [DOI] [PubMed] [Google Scholar]
- 35. Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 36. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Crystallography & NMR system. A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 37. Beavo J. A., Francis S. H., Houslay M. D. (2006) in Cyclic Nucleotide Phosphodiesterases in Health and Disease (Beavo J. A., Francis S. H., Houslay M. D., eds) CRC Press, Boca Raton, FL [Google Scholar]
- 38. Stevens J., Rambaut A. (2001) Evolutionary rate differences in trypanosomes. Infect. Genet. Evol. 1, 143–150 [DOI] [PubMed] [Google Scholar]
- 39. Nilsson D., Gunasekera K., Mani J., Osteras M., Farinelli L., Baerlocher L., Roditi I., Ochsenreiter T. (2010) Spliced leader trapping reveals widespread alternative splicing patterns in the highly dynamic transcriptome of Trypanosoma brucei. PLoS Pathog. 6, e1001037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kabani S., Fenn K., Ross A., Ivens A., Smith T. K., Ghazal P., Matthews K. (2009) Genome-wide expression profiling of in vivo-derived bloodstream parasite stages and dynamic analysis of mRNA alterations during synchronous differentiation in Trypanosoma brucei. BMC Genomics 10, 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jones A., Faldas A., Foucher A., Hunt E., Tait A., Wastling J. M., Turner C. M. (2006) Visualisation and analysis of proteomic data from the procyclic form of Trypanosoma brucei. Proteomics 6, 259–267 [DOI] [PubMed] [Google Scholar]
- 42. Ke H., Wang H. (2007) Crystal structures of phosphodiesterases and implications on substrate specificity and inhibitor selectivity. Curr. Top. Med. Chem. 7, 391–403 [DOI] [PubMed] [Google Scholar]
- 43. Wang H., Yan Z., Geng J., Kunz S., Seebeck T., Ke H. (2007) Crystal structure of the Leishmania major phosphodiesterase LmjPDEB1 and insight into the design of the parasite-selective inhibitors. Mol. Microbiol. 66, 1029–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Burgin A. B., Magnusson O. T., Singh J., Witte P., Staker B. L., Bjornsson J. M., Thorsteinsdottir M., Hrafnsdottir S., Hagen T., Kiselyov A. S., Stewart L. J., Gurney M. E. (2010) Design of phosphodiesterase 4D (PDE4D) allosteric modulators for enhancing cognition with improved safety. Nat. Biotechnol. 28, 63–70 [DOI] [PubMed] [Google Scholar]
- 45. Wang H., Liu Y., Huai Q., Cai J., Zoraghi R., Francis S. H., Corbin J. D., Robinson H., Xin Z., Lin G., Ke H. (2006) Multiple conformations of phosphodiesterase-5. Implications for enzyme function and drug development. J. Biol. Chem. 281, 21469–21479 [DOI] [PubMed] [Google Scholar]
- 46. Wang H., Ye M., Robinson H., Francis S. H., Ke H. (2008) Conformational variations of both phosphodiesterase-5 and inhibitors provide the structural basis for the physiological effects of vardenafil and sildenafil. Mol. Pharmacol. 73, 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S. (2002) The protein kinase complement of the human genome. Science 298, 1912–1934 [DOI] [PubMed] [Google Scholar]
- 48. Pils B., Schultz J. (2004) Evolution of the multifunctional protein tyrosine phosphatase family. Mol. Biol. Evol. 21, 625–631 [DOI] [PubMed] [Google Scholar]
- 49. Zettl M., Adrain C., Strisovsky K., Lastun V., Freeman M. (2011) Rhomboid family pseudoproteases use the ER quality control machinery to regulate intercellular signaling. Cell 145, 79–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jura N., Shan Y., Cao X., Shaw D. E., Kuriyan J. (2009) Structural analysis of the catalytically inactive kinase domain of the human EGF receptor 3. Proc. Natl. Acad. Sci. U.S.A. 106, 21608–21613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pils B., Schultz J. (2004) Inactive enzyme-homologues find new function in regulatory processes. J. Mol. Biol. 340, 399–404 [DOI] [PubMed] [Google Scholar]
- 52. Wang H., Yan Z., Yang S., Cai J., Robinson H., Ke H. (2008) Kinetic and structural studies of phosphodiesterase-8A and implication on the inhibitor selectivity. Biochemistry 47, 12760–12768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang H., Liu Y., Hou J., Zheng M., Robinson H., Ke H. (2007) Structural insight into substrate specificity of phosphodiesterase 10. Proc. Natl. Acad. Sci. U.S.A. 104, 5782–5787 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.