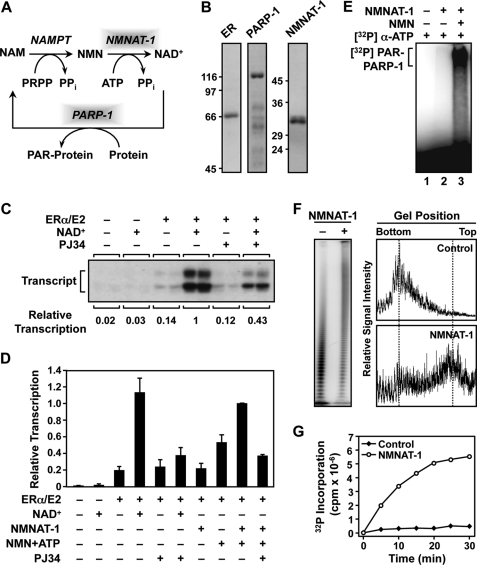
FIGURE 1.
NMNAT-1 produces NAD+ for PARP-1 automodification and PARP-1-dependent transcriptional regulation in vitro. A, the NAD+ salvage pathway produces NAD+ that is used by PARP-1 for PARylation of target proteins. NAM, nicotinamide; NAMPT, nicotinamide phosphoribosyltransferase; PRPP, phosphoribosyl pyrophosphate. B, Coomassie Blue-stained gels of purified recombinant human ERα, PARP-1, and NMNAT-1 used in the in vitro studies. C and D, NMNAT-1 enhances ERα-dependent transcription with chromatin templates through a PARP-dependent mechanism. Chromatin was assembled using Drosophila S190 extract and transcribed using HeLa cell nuclear extract. The reactions were carried out in the presence or absence of either 1) NAD+ (300 μm) or 2) NMNAT-1 (100 nm) with its substrates NMN and ATP (both at 300 μm). PJ34, an inhibitor of PARP enzymatic activity, was added as indicated. The bars shown in D represent the mean of at least four independent experiments, and the error bars represent S.E. E, NAD+ synthesized in vitro by NMNAT-1 can be used by PARP-1 for automodification. PARP-1 auto(ADP-ribosyl)ation assays were carried out using purified PARP-1 and NMNAT-1. [α-32P]ATP and NMN (both at 200 μm) were used as the substrates for NMNAT-1-dependent NAD+ synthesis. All reactions included sheared salmon sperm DNA as an activator of PARP-1 enzymatic activity. Automodified PARP-1 was analyzed by SDS-PAGE and autoradiography. F, NMNAT-1 stimulates PAR chain elongation by PARP-1 independently of NAD+ production. [32P]NAD+ (0.67 μm) was used as the substrate for the PARP-1 ADP-ribosylation assay. The reactions were carried out in the presence of 200 μm ATP but without NMN so NMNAT-1 was not able to produce NAD+. Left, the reactions were subjected to digestion with proteinase K to release free PAR, run on a 10% denaturing polyacrylamide gel to resolve the PAR chains of different lengths, and analyzed by a PhosphorImager. Right, the relative signal intensities of the PAR chains of different lengths were quantified by scanning along the middle of each sample lane. G, the total amount of PAR synthesized by PARP-1 is stimulated by NMNAT-1. PARP-1 automodification reactions were carried out using [32P]NAD+ (0.67 μm) and [α-32P]ATP (20 μm) at the same specific activity. The amount of radiolabel incorporated by PARP-1 into PAR chains in the presence or absence of NMNAT-1 was quantified by a filter binding assay over the course of a 30-min reaction.