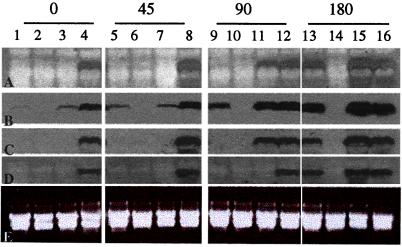
Figure 4.
Northern blot analysis of the induction or derepression of synthesis of mRNAs encoded by four different CO2-responsive genes on shift of cells from high concentrations of CO2 to low, ambient concentrations of CO2. Total RNA was extracted from wild-type cells (lanes 1, 5, 9, and 13), cia5 mutant cells (lanes 2, 6, 10, and 14), cia5 mutants complemented with an intact Cia5 gene construct (lanes 3, 7, 11, and 15), and cia5 mutants complemented with a truncated Cia5 gene (lanes 4, 8, 12, and 16). Total RNA was extracted from cells maintained in high concentrations of CO2 (lanes 1–4) or from cells after they were switched from high CO2 levels to low levels of CO2 for 45 min (lanes 5–8), 90 min (lanes 9–12), or 180 min (lanes 13–16). (A) mRNAs detected by hybridization with probes produced from the cDNA encoding Ccp2 (LIP36), a chloroplast inner envelope protein. (B) mRNAs detected with hybridization probes to the CAH1 cDNA encoding the major C. reinhardtii periplasmic CA. (C) mRNAs detected with probes to Att1 cDNA sequences encoding an alanine:α-ketoglutarate aminotransferase. (D) mRNAs detected with probes produced from the mtCA1 cDNA encoding a mitochondrial CA. (E) Ethidium bromide-stained gel illustrating uniform loading of total RNAs in each lane. The same RNA preparations used for the Northern blot analyses depicted in Fig. 1B were used for the blots shown.