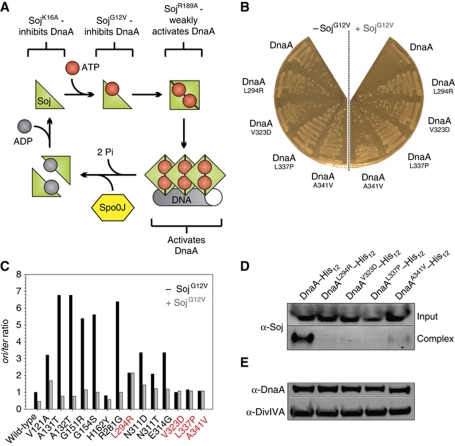
Figure 1.
Specific mutations in dnaA either bypass or suppress the inhibition of DNA replication initiation by SojG12V. (A) Pathway of the Soj activity cycle. (B) Point mutations in DnaA introduced by error-prone PCR were found to overcome the small colony phenotype characteristic of SojG12V overexpression. Strains were grown on NA plates in the presence or absence of 1% xylose to induce sojG12V expression. Wild-type (HM524), DnaAL294R (HM527), DnaAV323D (HM528), DnaAL337P (HM529), DnaAA341V (HM530). (C) The oriC-to-terminus ratios of dnaA point mutations generated using PCR mutagenesis were determined using MFA in the presence and absence of SojG12V overexpression (1% xylose). Suppressor mutations (red) were found to be recalcitrant to SojG12V activity. Cells were grown in LB medium at 30°C. Values were normalized to the ori:ter ratio of the wild-type strain grown in the absence of xylose. DnaAV121A (HM713), DnaAA131T (HM714), DnaAA132T (HM710), DnaAG151R (HM705), DnaAG154S (HM706), DnaAH162Y (HM707), DnaAR281G (HM708), DnaAN311D (HM712), DnaAN311T (HM709), DnaAE314G (HM711). (D) The SojG12V suppressor mutations in dnaA perturb the formation of a Soj:DnaA–His12 complex in vivo. Cells were grown in LB medium at 30°C, crosslinked with formaldehyde, and the DnaA–His12 complexes were purified before the crosslinks were reversed and proteins were separated by SDS–PAGE. Soj and DnaA–His12 were detected by western blot analysis. The top panel shows the amount of Soj protein in the cell lysate (Input) and the bottom panel shows the amount of Soj found in a complex with DnaA–His12 following purification (Complex). DnaA–His12 (HM657), DnaAL294R–His12 (HM716), DnaAV323D–His12 (HM555), DnaAL337P–His12 (HM658), DnaAA341V–His12 (HM725). (E) The amount of each DnaASup–His12 protein was determined by western blot analysis. DivIVA was used as a loading control.