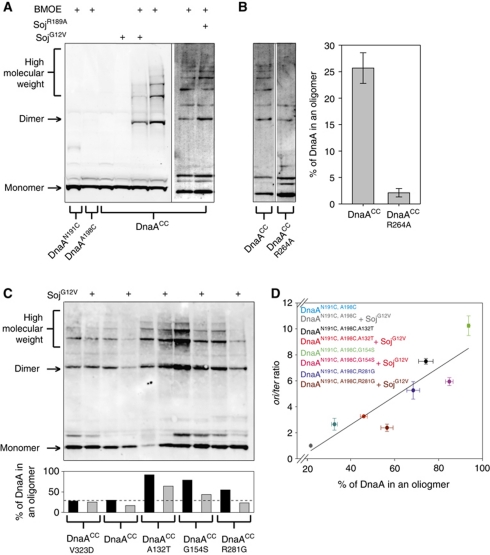
Figure 6.
The DnaA oligomer forms in vivo and is inhibited by monomeric Soj. (A) A DnaA oligomer can be isolated in vivo and is specifically inhibited by monomeric Soj. Cells were grown in crosslinking medium at 30°C to an A600 of 0.5, washed, and resuspended in in vivo crosslinking buffer in the presence of BMOE. Where indicated, 1% xylose was added or removed (in the case of the DnaAHyp strains) at an OD600 of 0.1 to modulate Soj protein expression. The identity of DnaA and/or Soj proteins are indicated below and above each lane, respectively. Proteins were separated by SDS–PAGE and DnaA was visualized by western blotting. DnaACC SojG12V (sGJS006), DnaAN198C (sGJS021), DnaAN191C (sGJS022), DnaACC SojR189A (sGJS033). (B) The DnaA oligomer is dependent on the arginine finger (R264) in vivo. Because the arginine finger mutation (dnaAR264A) is lethal (Duderstadt et al, 2010), the allele was introduced into a strain harbouring a plasmid origin (oriN) that allows viability in the absence of DnaA and/or oriC (Hassan et al, 1997). The identity of the DnaA proteins expressed are indicated below the respective lanes. DnaACC (sGJS051), DnaACC,R264A (sGJS052). Quantification of three biological repeats is shown. Error bars represent the standard deviation of the data. (C) The hypermorphic DnaA mutants have an increased propensity to form oligomers in vivo. Reaction conditions and figure annotations are as described in (A). The bar chart below shows the quantification of the gel. DnaA was defined as being in a helix if found in a dimer or higher molecular weight complex. DnaACC (sGJS006), DnaACC,V323D (sGJS008), DnaACC,A132T (sGJS036), DnaACC,G154S (sGJS037), DnaACC,R281G (sGJS038). (D) The percentage of DnaA found in an oligomer correlates with initiation frequency. The reactions in (C) (with the exception of DnaAV323D) were repeated in triplicate, the lanes quantified using ImageJ, and the percentage of DnaA found in the helix calculated as described above. Error bars represent the s.e.m. of the data. Concurrently, the oriC-to-terminus ratios were determined by MFA. Errors bars represent the standard deviation of the data. Linear regression analysis yields an R2 value of 0.82 while the Pearson product moment correlation coefficient is 0.903 (P-value=0.00212).