Figure 1.
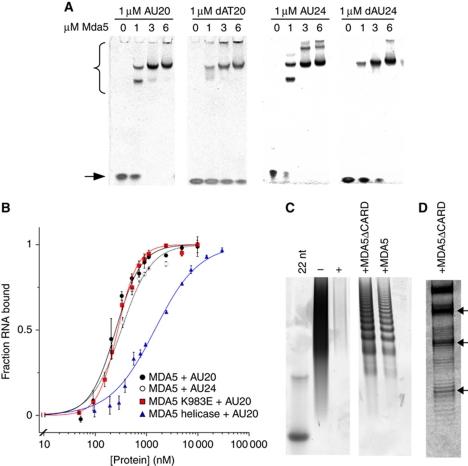
MDA5 binds short nucleic acid ligands with a 16–18-bp footprint. (A) Native PAGE showing the mobility shift of 20- and 24-bp RNA and DNA oligonucleotides (stained with SYBR Green) by full-length MDA5. MDA5–oligonucleotide complexes (bracket) migrate more slowly than free oligonucleotides (arrow). (B) Binding curves based on densitometry of bands in EMSA. MDA5 bound AU20 with Kd=287±13 nM and a Hill coefficient, n=1.7 (black squares). MDA5 bound AU24 with Kd=360±22 nM, n=1.5 (open squares). MDA5 K983E bound AU20 with Kd=277±14.8 nM, n=2.1 (red). The MDA5 helicase construct bound AU20 with Kd=1.45±0.05 μM, n=1 (blue). Each curve is the mean±s.e.m. of three to four experiments. (C) Poly(I:C) RNase protection with full-length and CARD-deleted MDA5 on denaturing PAGE. Both constructs protect poly(I:C) from RNases I and V1 in 16–18-bp increments with the smallest band at ∼50 nt. Poly(I:C) (− lane) is completely degraded by RNases (+ lane). Left lane, 22-bp DNA marker. (D) High-resolution gel showing the fine structure of bands in the RNase protection experiment from (C). See also Supplementary Figure S1.