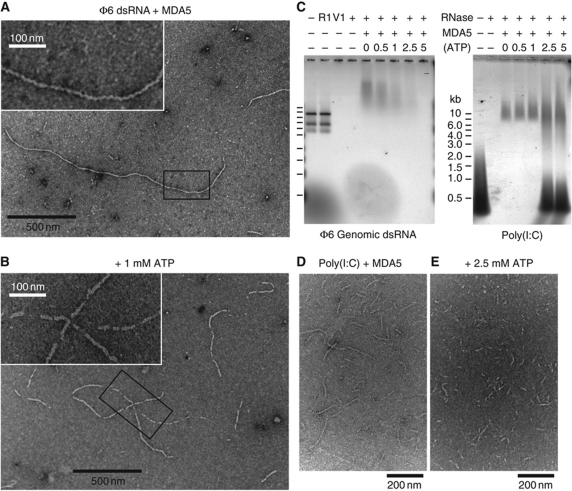
Figure 6.
MDA5 assembles into regular filaments on long dsRNA ligands. (A) Negative-stain electron micrograph (EM) of MDA5 mixed with bacteriophage ϕ6 genomic dsRNA shows filaments 8–10 nm in diameter. Filaments have few breaks along their length and can be up to 1.8 μm long. Inset: close-up of the boxed region. (B) 1 mM ATP induces breaks in the filaments. At 5 mM ATP, no filaments were observed. Inset: close-up of the boxed region. (C) Nuclease protection assay shows ϕ6 RNA is not protected by MDA5 when ATP is present (left). The three ϕ6 genome segments (2.9, 4.1 and 6.4 kb) are degraded by dsRNA endonuclease V1 (V1), but not ssRNA endonuclease 1 (R1). MDA5 reduces the mobility of ϕ6 and poly(I:C) RNA on agarose gel electrophoresis and protects them from RNases. Increasing ATP levels decrease RNase protection by MDA5 of ϕ6 RNA (left) but not of poly(I:C) RNA (right). At >2.5 mM ATP, free poly(I:C) is also protected. (D, E) EM of poly(I:C) and MDA5 without ATP (D), and with 2.5 mM ATP (E). Filaments incubated with ATP tend to aggregate, most likely from enhanced filament crosslinking. This effect is not observed with AMPPNP. See Supplementary Figure S6 for protein-only and RNA-only controls.