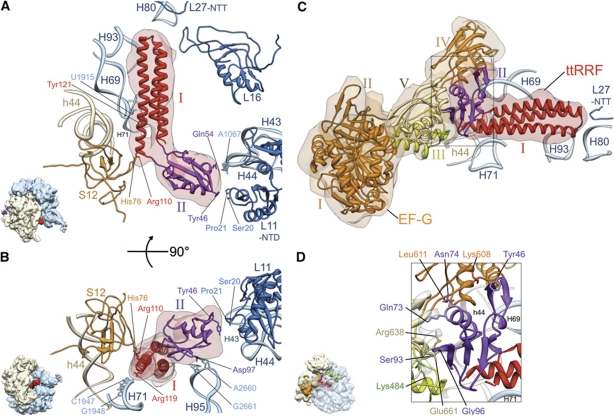
Figure 2.
Interactions between the ribosome and ttRRF in complex 1 (A, B), and between ttRRF and EF-G in complex 2 (C, D). (A) Flexibly fitted atomic structure of ttRRF (red and purple ribbons, PDB ID; 1EH1) into the corresponding RRF density (semitransparent pink). Contacts and proximities between the amino-acid residues of ttRRF and ribosomal components, such as amino-acid residues of proteins L11 and S12 and nucleotides of the 23S rRNA helices 43 (H43) and 69 (H69) are indicated. Domains I and II of the fitted ttRRF coordinates are depicted in red and purple, respectively. (B) Same as in (A), but rotated around a horizontal axis by ∼90°, to reveal interactions between an amino-acid residue of domain II of ttRRF and conserved nucleotides of the 23S rRNA helix 95 (H95), and between domain I of ttRRF and helix 71 (H71) of the 23S rRNA. (C) Flexibly fitted atomic structures of ttRRF and the homology model of E. coli EF-G into the corresponding cryo-EM densities of ttRRF (semitransparent red, in the intermediate position Pi) and EF-G (semitransparent orange), respectively, are shown with the ribosomal components present in the immediate vicinity of RRF. EF-G domains I–V are shown in distinctive colours, I (orange), II (brown), III (green), IV (orange) and V (yellow). L27-NTT refers to the N-terminal tail of protein L27. (D) Enlarged boxed area in (C) to reveal ribosomal neighbourhood of ttRRF in the position Pi and interaction of domain II of ttRRF with EF-G domains III–V. Five select pairs of amino-acid interactions are labelled (see also Supplementary Table S1B for the complete list of amino-acid interactions between ttRRF and EF-G). All helices of the 16S and 23S rRNAs are identified with h and H, respectively; while ribosomal proteins of the small and large subunits are prefixed by S and L, respectively. Thumbnails to the lower left of (A, B, D) depict overall orientations of the ribosome.