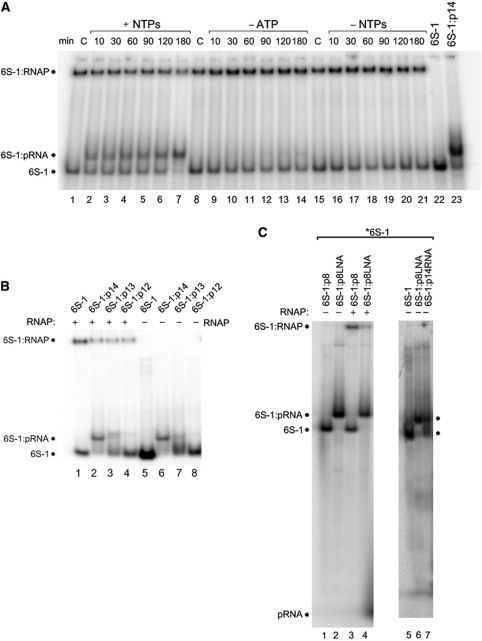
Figure 2.
Role of pRNA length and pRNA:6S-1 RNA duplex stability. (A) Trace amounts (<1 nM) of 5′-endlabelled 6S-1 RNA and 2.5 μM unlabelled 6S-1 RNA were subjected to the annealing procedure in a volume of 4 μl (see Materials and methods); then, 1 μl of a heparin solution (400 ng/ μl) and 2 μl 5 × activity buffer were added and samples were kept at 37°C; then 1.06 μl RNAP holoenzyme (8 μg/μl) were added and samples were incubated for 30 min at 37°C, followed by addition of 2 μl nucleotide solution (all four NTPs: lanes 2–7; only CTP, GTP and UTP: lanes 9–14) or 2 μl of double-distilled water instead (no NTPs; lanes 15–21) (final volume 10 μl; f.c. RNAP: 2 μM; f.c. 6S-1 RNA: 1 μM; f.c. NTPs: 200 μM each); lanes C: samples incubated for 180 min at 37°C in the absence of NTPs. After transcription at 37°C for the time period indicated above each lane, samples were analysed by 7.5% native PAGE (1 × TBE). (B) Trace amounts (<1 nM) of 5′-endlabelled 6S-1 RNA and 1.7 μM unlabelled 6S-1 RNA, either alone (lane 5) or in the presence of 17 μM pRNA 14-mer (lane 6), 13-mer (5′-GUU CGG UCA AAA C, lane 7) or 12-mer (5′-GUU CGG UCA AAA, lane 8) were annealed in 6 μl 1 × TE buffer and then loaded on a 7.5% native PAA gel (1 × TBE); lanes 1–4: as lanes 5–8, but before gel loading 1 μl of a heparin solution (400 ng/μl) and 2 μl 5 × activity buffer were added and samples were kept at 37°C; then 1.06 μl RNAP holoenzyme (8 μg/μl) was added and samples were incubated for 30 min at 37°C followed by gel loading. (C) Test for annealing of the pRNA 8-mer (lane 1) or an isosequential all-LNA 8-mer (lane 2) to 6S-1 RNA; lanes 3 and 4: as lanes 1 and 2, but after the annealing procedure samples were further incubated with RNAP holoenzyme; for details, see (B) and Materials and methods. Lanes 5–7 illustrate that 6S-1 RNA hybrid complexes with the LNA 8-mer cause the same mobility shift as obtained with the RNA 14-mer.