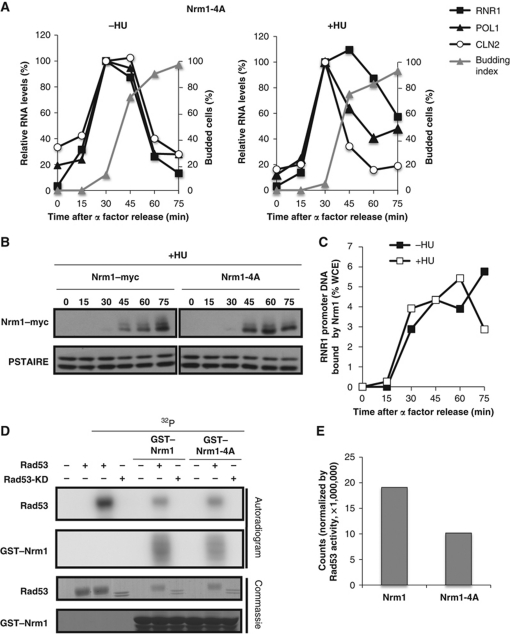
Figure 6.
Mutation of Rad53-dependent phosphorylation sites in Nrm1 impairs its function in response to DNA replication stress. (A) MBF-regulated transcription is not induced in response to HU in the Nrm1-4A mutant. Nrm1-4A mutant cells were grown in rich medium, synchronized by α-factor and subsequently released into media without (left panel) or with (right panel) 0.2 M HU. CLN2 (SBF-regulated) and RNR1 and POL1 (MBF-regulated) RNA from arrested cells (0 min) and cells released from the arrest for the indicated interval was quantitated by RT–qPCR and shown as percentage of the maximal RNA level in untreated cells. (B) Nrm1-4A is less phosphorylated in vivo in response to replication stress. Nrm1–myc and Nrm1-4A–myc were detected by immunoblot using cells from the same time course as in (A). Anti-PSTAIRE is shown as a loading control. (C) Nrm1-4A binds to MBF-dependent promoters despite the presence of HU. Nrm1-4A–myc binding to RNR1 promoter DNA was determined by ChIP in samples from the same time course as in (A). Values are shown as percentage of WCE. (D) Nrm1-4A is less phosphorylated than Nrm1 wild-type in vitro by Rad53. An in vitro kinase assay was performed using γ-32P-ATP and bacterially expressed recombinant Rad53 protein kinase or Rad53-KD and either recombinant GST–Nrm1 or GST–Nrm1-4A (second panel). Rad53 protein kinase activity is indicated by its autophosphorylation (top panel). Protein levels on the same gel are shown by Commassie Blue staining (lower two panels). (E) Quantification of Nrm1 and Nrm1-4A phosphorylation by Rad53 in the in vitro kinase assay shown in (D). Counts are normalized by Rad53 activity in each sample.